Estimating the basic reproduction number at the beginning of an outbreak
- PMID: 35714080
- PMCID: PMC9205483
- DOI: 10.1371/journal.pone.0269306
Estimating the basic reproduction number at the beginning of an outbreak
Abstract
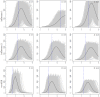
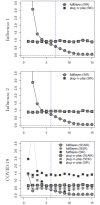
We compare several popular methods of estimating the basic reproduction number, R0, focusing on the early stages of an epidemic, and assuming weekly reports of new infecteds. We study the situation when data is generated by one of three standard epidemiological compartmental models: SIR, SEIR, and SEAIR; and examine the sensitivity of the estimators to the model structure. As some methods are developed assuming specific epidemiological models, our work adds a study of their performance in both a well-specified (data generating model and method model are the same) and miss-specified (data generating model and method model differ) settings. We also study R0 estimation using Canadian COVID-19 case report data. In this study we focus on examples of influenza and COVID-19, though the general approach is easily extendable to other scenarios. Our simulation study reveals that some estimation methods tend to work better than others, however, no singular best method was clearly detected. In the discussion, we provide recommendations for practitioners based on our results.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, et al.. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: A data-driven analysis in the early phase of the outbreak. International Journal of Infectious Diseases. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050 - DOI - PMC - PubMed
-
- Mellan TA, Hoeltgebaum HH, Mishra S, Whittaker C, Schnekenberg RP, Gandy A, et al.. Report 21: Estimating COVID-19 cases and reproduction number in Brazil. medRxiv. 2020.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

