Zika virus induces FOXG1 nuclear displacement and downregulation in human neural progenitors
- PMID: 35714598
- PMCID: PMC9287670
- DOI: 10.1016/j.stemcr.2022.05.008
Zika virus induces FOXG1 nuclear displacement and downregulation in human neural progenitors
Abstract
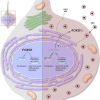
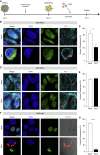
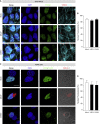
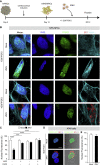
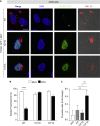
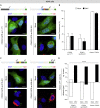
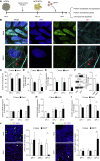
Congenital alterations in the levels of the transcription factor Forkhead box g1 (FOXG1) coding gene trigger "FOXG1 syndrome," a spectrum that recapitulates birth defects found in the "congenital Zika syndrome," such as microcephaly and other neurodevelopmental conditions. Here, we report that Zika virus (ZIKV) infection alters FOXG1 nuclear localization and causes its downregulation, thus impairing expression of genes involved in cell replication and apoptosis in several cell models, including human neural progenitor cells. Growth factors, such as EGF and FGF2, and Thr271 residue located in FOXG1 AKT domain, take part in the nuclear displacement and apoptosis protection, respectively. Finally, by progressive deletion of FOXG1 sequence, we identify the C-terminus and the residues 428-481 as critical domains. Collectively, our data suggest a causal mechanism by which ZIKV affects FOXG1, its target genes, cell cycle progression, and survival of human neural progenitors, thus contributing to microcephaly.
Keywords: FGF2; FOXG1; Zika virus; autism; brain cancer; congenital brain malformations; hiPSCs; microcephaly; neural stem cells; neurodevelopment; neurotropic virus.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Baek S.T., Copeland B., Yun E.-J., Kwon S.-K., Guemez-Gamboa A., Schaffer A.E., Kim S., Kang H.-C., Song S., Mathern G.W., et al. An AKT3-FOXG1-reelin network underlies defective migration in human focal malformations of cortical development. Nat. Med. 2015;21:1445–1454. doi: 10.1038/nm.3982. - DOI - PMC - PubMed
-
- Blackmon K., Waechter R., Landon B., Noël T., Macpherson C., Donald T., Cudjoe N., Evans R., Burgen K.S., Jayatilake P., et al. Epilepsy surveillance in normocephalic children with and without prenatal Zika virus exposure. PLoS Neglected Trop. Dis. 2020;14:e0008874. doi: 10.1371/journal.pntd.0008874. - DOI - PMC - PubMed
-
- Bulstrode H., Johnstone E., Marques-Torrejon M.A., Ferguson K.M., Bressan R.B., Blin C., Grant V., Gogolok S., Gangoso E., Gagrica S., et al. Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes Dev. 2017;31:757–773. doi: 10.1101/gad.293027.116. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

