Comparative parallel multi-omics analysis during the induction of pluripotent and trophectoderm states
- PMID: 35715410
- PMCID: PMC9205865
- DOI: 10.1038/s41467-022-31131-8
Comparative parallel multi-omics analysis during the induction of pluripotent and trophectoderm states
Abstract
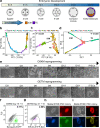
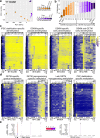
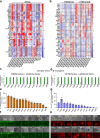
Following fertilization, it is only at the 32-64-cell stage when a clear segregation between cells of the inner cell mass and trophectoderm is observed, suggesting a 'T'-shaped model of specification. Here, we examine whether the acquisition of these two states in vitro, by nuclear reprogramming, share similar dynamics/trajectories. Using a comparative parallel multi-omics analysis (i.e., bulk RNA-seq, scRNA-seq, ATAC-seq, ChIP-seq, RRBS and CNVs) on cells undergoing reprogramming to pluripotency and TSC state we show that each reprogramming system exhibits specific trajectories from the onset of the process, suggesting 'V'-shaped model. We describe in detail the various trajectories toward the two states and illuminate reprogramming stage-specific markers, blockers, facilitators and TSC subpopulations. Finally, we show that while the acquisition of the TSC state involves the silencing of embryonic programs by DNA methylation, during the acquisition of pluripotency these regions are initially defined but retain inactive by the elimination of H3K27ac.
© 2022. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

