Selective cell cycle arrest in glioblastoma cell lines by quantum molecular resonance alone or in combination with temozolomide
- PMID: 35715634
- PMCID: PMC9427848
- DOI: 10.1038/s41416-022-01865-9
Selective cell cycle arrest in glioblastoma cell lines by quantum molecular resonance alone or in combination with temozolomide
Abstract
Background: Glioblastoma is the most aggressive form of brain cancer, characterised by high proliferation rates and cell invasiveness. Despite advances in surgery and radio-chemotherapy, patients continue to have poor prognoses, with a survival rate of 14-15 months. Thus, new therapeutic strategies are needed. Non-ionising electromagnetic fields represent an emerging option given the potential advantages of safety, low toxicity and the possibility to be combined with other therapies.
Methods: Here, the anticancer activity of quantum molecular resonance (QMR) was investigated. For this purpose, three glioblastoma cell lines were tested, and the QMR effect was evaluated on cancer cell proliferation rate and aggressiveness. To clarify the QMR mechanism of action, the proteomic asset after stimulation was delineated. Mesenchymal stromal cells and astrocytes were used as healthy controls.
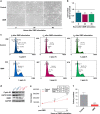
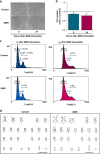
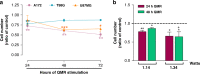
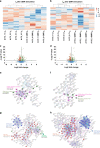
Results: QMR affected cancer cell proliferation, inducing a significant arrest of cell cycle progression and reducing cancer tumorigenicity. These parameters were not altered in healthy control cells. Proteomic analysis suggested that QMR acts not only on DNA replication but also on the machinery involved in the mitotic spindle assembly and chromosome segregation. Moreover, in a combined therapy assessment, QMR significantly enhanced temozolomide efficacy.
Conclusions: QMR technology appears to be a promising tool for glioblastoma treatment.
© 2022. The Author(s).
Conflict of interest statement
DC, GM, AB, MB, KC, MM, AM, PC, RB, DP, FAR, LP, MR and GA declare no competing interests. GP is the cofounder and AP is an employee of Telea Electronic Engineering srl, Sandrigo, Italy. They had no role in the design, execution, interpretation or writing of the study.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

