Endothelial and hematopoietic hPSCs differentiation via a hematoendothelial progenitor
- PMID: 35715824
- PMCID: PMC9205076
- DOI: 10.1186/s13287-022-02925-w
Endothelial and hematopoietic hPSCs differentiation via a hematoendothelial progenitor
Abstract
Background: hPSC-derived endothelial and hematopoietic cells (ECs and HCs) are an interesting source of cells for tissue engineering. Despite their close spatial and temporal embryonic development, current hPSC differentiation protocols are specialized in only one of these lineages. In this study, we generated a hematoendothelial population that could be further differentiated in vitro to both lineages.
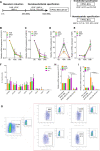
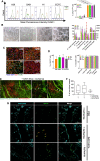
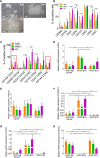
Methods: Two hESCs and one hiPSC lines were differentiated into a hematoendothelial population, hPSC-ECs and blast colonies (hPSC-BCs) via CD144+-embryoid bodies (hPSC-EBs). hPSC-ECs were characterized by endothelial colony-forming assay, LDL uptake assay, endothelial activation by TNF-α, nitric oxide detection and Matrigel-based tube formation. Hematopoietic colony-forming cell assay was performed from hPSC-BCs. Interestingly, we identified a hPSC-BC population characterized by the expression of both CD144 and CD45. hPSC-ECs and hPSC-BCs were analyzed by flow cytometry and RT-qPCR; in vivo experiments have been realized by ischemic tissue injury model on a mouse dorsal skinfold chamber and hematopoietic reconstitution in irradiated immunosuppressed mouse from hPSC-ECs and hPSC-EB-CD144+, respectively. Transcriptomic analyses were performed to confirm the endothelial and hematopoietic identity of hESC-derived cell populations by comparing them against undifferentiated hESC, among each other's (e.g. hPSC-ECs vs. hPSC-EB-CD144+) and against human embryonic liver (EL) endothelial, hematoendothelial and hematopoietic cell subpopulations.
Results: A hematoendothelial population was obtained after 84 h of hPSC-EBs formation under serum-free conditions and isolated based on CD144 expression. Intrafemorally injection of hPSC-EB-CD144+ contributed to the generation of CD45+ human cells in immunodeficient mice suggesting the existence of hemogenic ECs within hPSC-EB-CD144+. Endothelial differentiation of hPSC-EB-CD144+ yields a population of > 95% functional ECs in vitro. hPSC-ECs derived through this protocol participated at the formation of new vessels in vivo in a mouse ischemia model. In vitro, hematopoietic differentiation of hPSC-EB-CD144+ generated an intermediate population of > 90% CD43+ hPSC-BCs capable to generate myeloid and erythroid colonies. Finally, the transcriptomic analyses confirmed the hematoendothelial, endothelial and hematopoietic identity of hPSC-EB-CD144+, hPSC-ECs and hPSC-BCs, respectively, and the similarities between hPSC-BC-CD144+CD45+, a subpopulation of hPSC-BCs, and human EL hematopoietic stem cells/hematopoietic progenitors.
Conclusion: The present work reports a hPSC differentiation protocol into functional hematopoietic and endothelial cells through a hematoendothelial population. Both lineages were proven to display characteristics of physiological human cells, and therefore, they represent an interesting rapid source of cells for future cell therapy and tissue engineering.
Keywords: Endothelial differentiation; Endothelium; Hematopoietic cells; Hematopoietic differentiation; Hemogenic endothelium; hESCs; hPSC; hiPSC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests in relation to this study.
Figures



References
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. Available from http://www.ncbi.nlm.nih.gov/pubmed/9804556. - PubMed
-
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (80-). 2007 Dec 21;318(5858):1917 LP—1920. Available from http://science.sciencemag.org/content/318/5858/1917.abstract. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

