Exome sequencing identifies genetic variants in anophthalmia and microphthalmia
- PMID: 35716026
- PMCID: PMC9283271
- DOI: 10.1002/ajmg.a.62874
Exome sequencing identifies genetic variants in anophthalmia and microphthalmia
Abstract
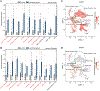
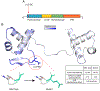
Anophthalmia and microphthalmia (A/M) are rare birth defects affecting up to 2 per 10,000 live births. These conditions are manifested by the absence of an eye or reduced eye volumes within the orbit leading to vision loss. Although clinical case series suggest a strong genetic component in A/M, few systematic investigations have been conducted on potential genetic contributions owing to low population prevalence. To overcome this challenge, we utilized DNA samples and data collected as part of the National Birth Defects Prevention Study (NBDPS). The NBDPS employed multi-center ascertainment of infants affected by A/M. We performed exome sequencing on 67 family trios and identified numerous genes affected by rare deleterious nonsense and missense variants in this cohort, including de novo variants. We identified 9 nonsense changes and 86 missense variants that are absent from the reference human population (Genome Aggregation Database), and we suggest that these are high priority candidate genes for A/M. We also performed literature curation, single cell transcriptome comparisons, and molecular pathway analysis on the candidate genes and performed protein structure modeling to determine the potential pathogenic variant consequences on PAX6 in this disease.
Keywords: congenital abnormalities; genetic epidemiology; newborn eye abnormalities.
© 2022 Wiley Periodicals LLC.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

