Ligand-based and structure-based studies to develop predictive models for SARS-CoV-2 main protease inhibitors through the 3d-qsar.com portal
- PMID: 35716228
- PMCID: PMC9206107
- DOI: 10.1007/s10822-022-00460-7
Ligand-based and structure-based studies to develop predictive models for SARS-CoV-2 main protease inhibitors through the 3d-qsar.com portal
Abstract
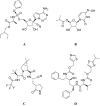
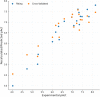
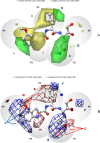
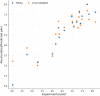
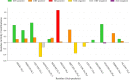
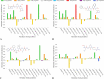
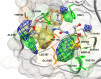
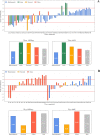
The main protease (Mpro) of SARS-Cov-2 is the essential enzyme for maturation of functional proteins implicated in viral replication and transcription. The peculiarity of its specific cleavage site joint with its high degree of conservation among all coronaviruses promote it as an attractive target to develop broad-spectrum inhibitors, with high selectivity and tolerable safety profile. Herein is reported a combination of three-dimensional quantitative structure-activity relationships (3-D QSAR) and comparative molecular binding energy (COMBINE) analysis to build robust and predictive ligand-based and structure-based statistical models, respectively. Models were trained on experimental binding poses of co-crystallized Mpro-inhibitors and validated on available literature data. By means of deep optimization both models' goodness and robustness reached final statistical values of r2/q2 values of 0.97/0.79 and 0.93/0.79 for the 3-D QSAR and COMBINE approaches respectively, and an overall predictiveness values of 0.68 and 0.57 for the SDEPPRED and AAEP metrics after application to a test set of 60 compounds covered by the training set applicability domain. Despite the different nature (ligand-based and structure-based) of the employed methods, their outcome fully converged. Furthermore, joint ligand- and structure-based structure-activity relationships were found in good agreement with nirmatrelvir chemical features properties, a novel oral Mpro-inhibitor that has recently received U.S. FDA emergency use authorization (EUA) for the oral treatment of mild-to-moderate COVID-19 infected patients. The obtained results will guide future rational design and/or virtual screening campaigns with the aim of discovering new potential anti-coronavirus lead candidates, minimizing both time and financial resources. Moreover, as most of calculation were performed through the well-established web portal 3d-qsar.com the results confirm the portal as a useful tool for drug design.
Keywords: 3-D QSAR; COMBINE; Ligand-based drug design; SARS-Cov-2; Structure-activity relationships; Structure-based drug design.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- World Health Organization. WHO Director-General's remarks at the media briefing on 2019-nCoV on 2020 11 February 2020; Available from: https://www.who.int/director-general/speeches/detail/who-director-genera....
-
- World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020; Available from: https://www.who.int/director-general/speeches/detail/who-director-genera....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

