Potential disease-modifying therapies for Huntington's disease: lessons learned and future opportunities
- PMID: 35716694
- PMCID: PMC7613206
- DOI: 10.1016/S1474-4422(22)00121-1
Potential disease-modifying therapies for Huntington's disease: lessons learned and future opportunities
Abstract
Huntington's disease is the most frequent autosomal dominant neurodegenerative disorder; however, no disease-modifying interventions are available for patients with this disease. The molecular pathogenesis of Huntington's disease is complex, with toxicity that arises from full-length expanded huntingtin and N-terminal fragments of huntingtin, which are both prone to misfolding due to proteolysis; aberrant intron-1 splicing of the HTT gene; and somatic expansion of the CAG repeat in the HTT gene. Potential interventions for Huntington's disease include therapies targeting huntingtin DNA and RNA, clearance of huntingtin protein, DNA repair pathways, and other treatment strategies targeting inflammation and cell replacement. The early termination of trials of the antisense oligonucleotide tominersen suggest that it is time to reflect on lessons learned, where the field stands now, and the challenges and opportunities for the future.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests SJT receives research grant funding from the CHDI Foundation, Vertex Pharmaceuticals, the UK Medical Research Council, the Wellcome Trust (200181/Z/15/Z), and the UK Dementia Research Institute, which receives its funding from the UK Dementia Research Institute. The UK Dementia Research Institute is funded by the UK Medical Research Council, Alzheimer's Society, and Alzheimer's Research UK. SJT has undertaken consultancy services for Annexon, Alphasights, Alnylam Pharmaceuticals, Atalanta Therapeutics, Roche, Genentech, Guidepoint, Horama, Locanobio, LoQus23 Therapeutics, Novartis Pharma, PTC Therapeutics, Sanofi, Spark Therapeutics, Takeda Pharmaceuticals, Triplet Therapeutics, University College Irvine, Vertex Pharmaceuticals, and Wave Life Sciences. All honoraria for these consultancies were paid through the offices of University College London (UCL) Consultants, a wholly owned subsidiary of UCL. SJT is on the scientific advisory boards of Atalanta Therapeutics, LoQus 23 Therapeutics, and Triplet Therapeutics. SJT has a patent application number 2105484.6 on the FAN1-MLH1 interaction and structural analogues licensed to Adrestia Therapeutics. WRM is employed by the Leids Universitair Medisch Centrum (LUMC), which has patents on exon skipping approaches for neurological disorders, some of which have been licensed to ProQR and Amylon. Remunerations are paid to LUMC. WMCvR-M is an ad hoc consultant for Bridge Bio, has received project funding from Amylon and ProQR, and has received funding for contract research from UniQure. WMCvR-M is project leader in a project to develop an ASO therapy for SCA1 together with Vico Therapeutics. EJW reports grants from the CHDI Foundation, and Roche. EJW has undertaken consultancy work and advisory board work with Roche, Triplet Therapeutics, PTC Therapeutics, Takeda, Vico Therapeutics, Voyager, Huntington Study Group, Teitur Trophics, EcoR1 Capital, PTC Therapeutics, Annexon Biosciences, and Remix Therapeutics. All honoraria for these consultancies were paid through the offices of UCL Consultants. EJW holds a stock option for Triplet Therapeutics in part compensation for advisory board membership. AER is chair of the executive committee of the European Huntington Disease Network, which is funded by CHDI Foundation, with remuneration paid to Cardiff University. AER is also European co-principal investigator for the PROOF-HD trial, with remuneration paid to Cardiff University. AER also serves on the scientific advisory boards of Roche, Wave Pharma, Prilenia, and Triplet Therapeutics. AER has obtained research grants from the UK Medical Research Council, Health and Care Research Wales, EU Horizon 2020, Jacques and Gloria Gossweiler foundation, and is a chair of the Guarantors of Brain charity. BRL is on the scientific advisory board of sRNAlytics (GateHouse Bio), for which he received stock options, and reports scientific consultancy fees from Teva, Roche, Takeda, Triplet, UniQure, Novartis, Spark, Scintetica, LifeEdit, Design, Remix Therapeutics, and PTC Therapeutics. BRL's laboratory has obtained previous and current research grants from the Canadian Institutes of Health Research, Health Sciences Centre, Nanomedicines Innovation Network, CHDI Foundation, Teva, ProMIS, and uniQure. BRL is a founding co-editor-in-chief for the Journal of Huntington's Disease, former co-chair of the Huntington Study Group, and is co-founder and CEO of Incisive Genetics, in which he has stock and stock options (Incisive Genetics is an early-stage pre-clinical biotechnology company that was founded to develop in vivo lipid nanoparticle delivery of clustered regularly interspaced short palindromic repeats-Cas9 genome editing. This is not a therapeutic approach that is currently in clinical testing for Huntington's disease, nor is this approach in late pre-clinical stages. The company has no products to endorse, does not have an investigational new drug for Huntington's disease, and does not have any commercial efforts currently underway). CS serves as chief medical officer for CHDI Management and IM-S serves as vice president of Translational Biology for CHDI Management. CHDI Management is an advisor to CHDI Foundation. CHDI Foundation provided financial and scientific support to the Huntington's disease drug discovery programme developed by Ionis Pharmaceuticals through a development collaboration with Ionis Pharmaceuticals. Over time, CHDI Foundation was reimbursed for its financial support of the programme developed by Ionis Pharmaceuticals out of milestone payments received by Ionis Pharmaceuticals from Roche for progress in Roche's tominersen programme. CS received consultancy honoraria (unrelated to Huntington's disease programmes) from the following companies: Kyowa Kirin, Neuraly, the US Food and Drug Administration (New Drug Application), Neuroderm, Pinteon Pharmaceuticals, and vTv Therapeutics. CE-F receives support from a Wellcome Trust Collaborative Award (200181/Z/15/Z). Within the past 36 months MDF has received grant funding towards his research from the CHDI Foundation, the UK Dementia Research Institute, and the Rosetrees Trust. RIS reports no competing interests.
Figures


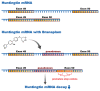
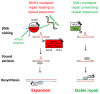
Comment in
-
Huntington's disease: disappointments and new beginnings.Lancet Neurol. 2022 Jul;21(7):582-584. doi: 10.1016/S1474-4422(22)00189-2. Lancet Neurol. 2022. PMID: 35716683 No abstract available.
References
-
- Leavitt BR, Kordasiewicz HB, Schobel SA. Huntingtin-Lowering Therapies for Huntington Disease. JAMA Neurol. 2020;77:764. - PubMed
-
- Bates GP, Dorsey R, Gusella JF, et al. Huntington disease. Nat Rev Dis Prim. 2015;1:1–21. - PubMed
-
- Tabrizi SJ, Ghosh R, Leavitt BR. Huntingtin Lowering Strategies for Disease Modification in Huntington’s Disease. Neuron. 2019;101:801–19. - PubMed
-
- Rüb U, Vonsattel JPV, Heinsen H, Korf H-W. The Neuropathology of Huntington’s disease: classical findings, recent developments and correlation to functional neuroanatomy. Adv Anat Embryol Cell Biol. 2015;217:1–146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

