The ecology of palm genomes: repeat-associated genome size expansion is constrained by aridity
- PMID: 35717562
- PMCID: PMC9796251
- DOI: 10.1111/nph.18323
The ecology of palm genomes: repeat-associated genome size expansion is constrained by aridity
Abstract
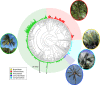
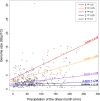
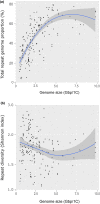
Genome size varies 2400-fold across plants, influencing their evolution through changes in cell size and cell division rates which impact plants' environmental stress tolerance. Repetitive element expansion explains much genome size diversity, and the processes structuring repeat 'communities' are analogous to those structuring ecological communities. However, which environmental stressors influence repeat community dynamics has not yet been examined from an ecological perspective. We measured genome size and leveraged climatic data for 91% of genera within the ecologically diverse palm family (Arecaceae). We then generated genomic repeat profiles for 141 palm species, and analysed repeats using phylogenetically informed linear models to explore relationships between repeat dynamics and environmental factors. We show that palm genome size and repeat 'community' composition are best explained by aridity. Specifically, Ty3-gypsy and TIR elements were more abundant in palm species from wetter environments, which generally had larger genomes, suggesting amplification. By contrast, Ty1-copia and LINE elements were more abundant in drier environments. Our results suggest that water stress inhibits repeat expansion through selection on upper genome size limits. However, elements that may associate with stress-response genes (e.g. Ty1-copia) have amplified in arid-adapted palm species. Overall, we provide novel evidence of climate influencing the assembly of repeat 'communities'.
Keywords: Arecaceae (palms); adaptation; ecology; genome size; phylogenetic regression; plant evolution; trait evolution; transposable elements.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures

References
-
- Andrews S. 2010. Fastqc: a quality control tool for high throughput sequence data. v.0.11.9 . Cambridge, UK. [WWW document] URL http://www.bioinformatics.babraham.ac.uk/projects/fastqc [accessed 1 January 2020].
-
- Baker WJ, Dransfield J. 2016. Beyond genera Palmarum: progress and prospects in palm systematics. Botanical Journal of the Linnean Society 182: 207–233.
-
- Barton K, Barton MK. 2015. Package ‘mumin’. v.1 . [WWW document] URL https://cran.r‐project.org/web/packages/MuMIn/index.html [accessed 1 January 2020].
-
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. 2008. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist 179: 975–986. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

