CRISPR/Cas12a-Based Diagnostic Platform Accurately Detects Nocardia farcinica Targeting a Novel Species-Specific Gene
- PMID: 35719360
- PMCID: PMC9198645
- DOI: 10.3389/fcimb.2022.884411
CRISPR/Cas12a-Based Diagnostic Platform Accurately Detects Nocardia farcinica Targeting a Novel Species-Specific Gene
Abstract
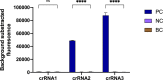
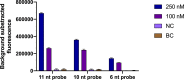
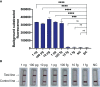
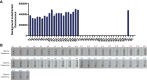
Under the COVID-19 pandemic background, nucleic acid detection has become the gold standard to rapidly diagnose the infectious disease. A rapid, low cost, reliable nucleic acid detection platform will be the key to control next potential pandemic. In this study, a nucleic acid detection platform, which combined CRISPR/Cas12a-based detection with loop-mediated isothermal amplification (LAMP), was developed and termed CRISPR-CLA. In the CRISPR-CLA system, LAMP preamplification was employed, and CRISPR/Cas12a-based detection was used to monitor the preamplicons. The forward inner primer (FIP) was engineered with a protospacer adjacent motif (PAM) site TTTA of Cas12a effector at the linker region; thus, the CRISPR-CLA platform can detect any sequence as long as the primer design meets the requirement of LAMP. To demonstrate the validity of the CRISPR-CLA system, it was applied for the molecular diagnosis of nocardiosis caused by Nocardia farcinica (N. farcinica). A highly conserved and species-specific gene pbr1 of N. farcinica, which was first reported in this study, was used as the target of detection. A set of LAMP primers targeting a fragment of pbr1 of the N. farcinica reference strain IFM 10152 was designed according to the principle of CRISPR-CLA. Three CRISPR RNAs (crRNAs) with different lengths were designed, and the most efficient crRNA was screened out. Additionally, three single-strand DNA (ssDNA) probes were tested to further optimize the detection system. As a result, the N. farcinica CRISPR-CLA assay was established, and the whole detection process, including DNA extraction (20 min), LAMP preamplification (70°C, 40 min), and CRISPR/Cas12a-mediated detection (37°C, 8 min), can be completed within 70 min. A fluorescence reader (for fluorescence CRISPR-CLA) or a lateral flow biosensor (for lateral-flow CRISPR-CLA) can be the media of the result readout. Up to 132 strains were used to examine the specificity of N. farcinica CRISPR-CLA assay, and no cross-reaction was observed with non-N. farcinica templates. The limit of detection (LoD) of the N. farcinica CRISPR-CLA assay was 100 fg double-strand DNA per reaction. N. farcinica was detected accurately in 41 sputum specimens using the N. farcinica CRISPR-CLA assay, which showed higher specificity than a real-time qPCR method. Hence, the N. farcinica CRISPR-CLA assay is a rapid, economic and accurate method to diagnose N. farcinica infection.
Keywords: CRISPR; CRISPR-CLA; Cas12a; Nocardia farcinica; accurate diagnosis; nucleic acid detection; pbr1.
Copyright © 2022 Qiu, Xu, Liu, Ren, Han and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
A CRISPR-based nucleic acid detection platform (CRISPR-CPA): Application for detection of Nocardia farcinica.J Appl Microbiol. 2022 May;132(5):3685-3693. doi: 10.1111/jam.15424. Epub 2022 Feb 6. J Appl Microbiol. 2022. PMID: 34936163
-
LAMP-CRISPR-Cas12-based diagnostic platform for detection of Mycobacterium tuberculosis complex using real-time fluorescence or lateral flow test.Mikrochim Acta. 2021 Sep 20;188(10):347. doi: 10.1007/s00604-021-04985-w. Mikrochim Acta. 2021. PMID: 34542728
-
Cas12a/Guide RNA-Based Platforms for Rapidly and Accurately Identifying Staphylococcus aureus and Methicillin-Resistant S. aureus.Microbiol Spectr. 2023 Mar 21;11(2):e0487022. doi: 10.1128/spectrum.04870-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36943040 Free PMC article.
-
Non-canonical CRISPR/Cas12a-based technology: A novel horizon for biosensing in nucleic acid detection.Talanta. 2024 May 1;271:125663. doi: 10.1016/j.talanta.2024.125663. Epub 2024 Jan 16. Talanta. 2024. PMID: 38232570 Review.
-
CRISPR/Cas12a-based technology: A powerful tool for biosensing in food safety.Trends Food Sci Technol. 2022 Apr;122:211-222. doi: 10.1016/j.tifs.2022.02.030. Epub 2022 Mar 1. Trends Food Sci Technol. 2022. PMID: 35250172 Free PMC article. Review.
Cited by
-
Accurate, Sensitive, and Rapid Detection of Pseudomonas aeruginosa Based on CRISPR/Cas12b with One Fluid-Handling Step.Microbiol Spectr. 2023 Feb 14;11(1):e0352322. doi: 10.1128/spectrum.03523-22. Epub 2023 Jan 9. Microbiol Spectr. 2023. PMID: 36622174 Free PMC article.
-
Application of pre-amplification-based CRISPR-Cas nanostructured biosensors for bacterial detection.Nanomedicine (Lond). 2025 Apr;20(8):903-915. doi: 10.1080/17435889.2025.2476384. Epub 2025 Mar 7. Nanomedicine (Lond). 2025. PMID: 40052226 Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

