Genomic Characterization of Two Escherichia fergusonii Isolates Harboring mcr-1 Gene From Farm Environment
- PMID: 35719362
- PMCID: PMC9204285
- DOI: 10.3389/fcimb.2022.774494
Genomic Characterization of Two Escherichia fergusonii Isolates Harboring mcr-1 Gene From Farm Environment
Abstract
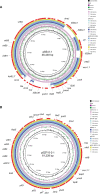
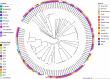
The prevalence and transmission of mobile colistin resistance (mcr) genes have led to a severe threat to humans and animals. Escherichia fergusonii is an emerging pathogen which is closely related to a variety of diseases. However, the report of mcr genes harboring E. fergusonii is still rare. One study in Brazil reported the E. fergusonii isolates with IncHI2-type plasmids harboring mcr-1. A Chinese study reported two strains carrying mcr-1 gene with the same plasmid type IncI2. Here, we identified two strains of E. fergusonii carrying mcr-1 gene from farm environments with IncX4-type and IncI2-type plasmids, respectively. To our best knowledge, this is the first report about mcr-1 gene located on IncX4-type plasmid in E. fergusonii. We investigate the resistance mechanism of colistin-resistant Escherichia fergusonii strains 6S41-1 and 5ZF15-2-1 and elucidate the genetic context of plasmids carrying mcr-1 genes. In addition, we also investigated chromosomal mutations mediated colistin resistance in these two strains. Species identification was performed using MALDI-TOF MS and 16S rRNA gene sequencing. The detection of mcr-1 gene was determined by PCR and Sanger sequencing. S1-pulsed-field gel electrophoresis (PFGE), Southern blotting, antimicrobial susceptibility testing, conjugation experiments, complete genome sequencing, and core genome analysis were conducted to investigate the characteristics of isolates harboring mcr-1. The mcr-1 genes on two strains were both plasmids encoded and the typical IS26-parA-mcr-1-pap2 cassette was identified in p6S41-1 while a nikA-nikB-mcr-1 locus sites on the conjugative plasmid p5ZF15-2-1. In addition, Core genome analysis reveals that E. fergusonii 6S41-1 and 5ZF15-2-1 have close genetic relationships. The mcr-1 gene is located on conjugative IncI2-type plasmid p5ZF15-2-1, which provides support for its further transmission. In addition, there's the possibility of mcr-1 spreading to humans through farm environments and thereby threatening public health. Therefore, continuous monitoring and investigations of mcr-1 among Enterobacteriaceae in farm environments are necessary to control the spread.
Keywords: IncI2; IncX4; core genome analysis; farm environments; whole-genome sequencing.
Copyright © 2022 Liu, Xu, Guo, Liu, Qiao, Ge, Zheng and Gou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Asai T., Kojima A., Harada K., Ishihara K., Takahashi T., Tamura Y. (2005). Correlation Between the Usage Volume of Veterinary Therapeutic Antimicrobials and Resistance in Escherichia Coli Isolated From the Feces of Food-Producing Animals in Japan. Jpn J. Infect. Dis. 58, 369–372. doi: 10.1097/01.qai.0000184861.26733.8c - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

