Novel Allosteric Mechanism of Dual p53/MDM2 and p53/MDM4 Inhibition by a Small Molecule
- PMID: 35720128
- PMCID: PMC9198586
- DOI: 10.3389/fmolb.2022.823195
Novel Allosteric Mechanism of Dual p53/MDM2 and p53/MDM4 Inhibition by a Small Molecule
Abstract
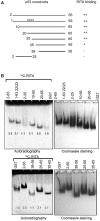
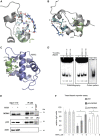
Restoration of the p53 tumor suppressor for personalised cancer therapy is a promising treatment strategy. However, several high-affinity MDM2 inhibitors have shown substantial side effects in clinical trials. Thus, elucidation of the molecular mechanisms of action of p53 reactivating molecules with alternative functional principle is of the utmost importance. Here, we report a discovery of a novel allosteric mechanism of p53 reactivation through targeting the p53 N-terminus which promotes inhibition of both p53/MDM2 (murine double minute 2) and p53/MDM4 interactions. Using biochemical assays and molecular docking, we identified the binding site of two p53 reactivating molecules, RITA (reactivation of p53 and induction of tumor cell apoptosis) and protoporphyrin IX (PpIX). Ion mobility-mass spectrometry revealed that the binding of RITA to serine 33 and serine 37 is responsible for inducing the allosteric shift in p53, which shields the MDM2 binding residues of p53 and prevents its interactions with MDM2 and MDM4. Our results point to an alternative mechanism of blocking p53 interaction with MDM2 and MDM4 and may pave the way for the development of novel allosteric inhibitors of p53/MDM2 and p53/MDM4 interactions.
Keywords: MDM2; MDMX(4); N-terminus; allosteric inhibition; p53.
Copyright © 2022 Grinkevich, Vema, Fawkner, Issaeva, Andreotti, Dickinson, Hedström, Spinnler, Inga, Larsson, Karlén, Wilhelm, Barran, Okorokov, Selivanova and Zawacka-Pankau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

