Identification of a neural development gene expression signature in colon cancer stem cells reveals a role for EGR2 in tumorigenesis
- PMID: 35720265
- PMCID: PMC9204726
- DOI: 10.1016/j.isci.2022.104498
Identification of a neural development gene expression signature in colon cancer stem cells reveals a role for EGR2 in tumorigenesis
Abstract
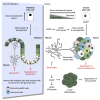
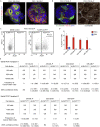
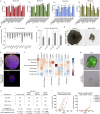
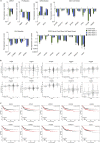
Recent evidence demonstrates that colon cancer stem cells (CSCs) can generate neurons that synapse with tumor innervating fibers required for tumorigenesis and disease progression. Greater understanding of the mechanisms that regulate CSC driven tumor neurogenesis may therefore lead to more effective treatments. RNA-sequencing analyses of ALDHPositive CSCs from colon cancer patient-derived organoids (PDOs) and xenografts (PDXs) showed CSCs to be enriched for neural development genes. Functional analyses of genes differentially expressed in CSCs from PDO and PDX models demonstrated the neural crest stem cell (NCSC) regulator EGR2 to be required for tumor growth and to control expression of homebox superfamily embryonic master transcriptional regulator HOX genes and the neural stem cell and master cell fate regulator SOX2. These data support CSCs as the source of tumor neurogenesis and suggest that targeting EGR2 may provide a therapeutic differentiation strategy to eliminate CSCs and block nervous system driven disease progression.
Keywords: Cancer; Stem cells research; Transcriptomics.
© 2022 The Authors.
Conflict of interest statement
A.S., T.J., D.M., and. D.H. are employees of Bayer AG. R.L., J.T., and M.L. are employees of Nuvisan ICB GmbH. C.R.A.R. is the founder of CELLphenomics GmbH.
Figures

References
-
- Adams J.C., Tucker R.P. The thrombospondin type 1 repeat (TSR) superfamily: diverse proteins with related roles in neuronal development. Dev. Dyn. 2000;218:280–299. - PubMed
LinkOut - more resources
Full Text Sources

