Evaluation of Methods for the Extraction of Spatial Muscle Synergies
- PMID: 35720729
- PMCID: PMC9202610
- DOI: 10.3389/fnins.2022.732156
Evaluation of Methods for the Extraction of Spatial Muscle Synergies
Abstract
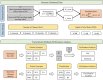
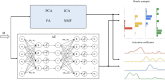
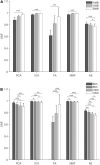
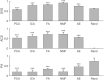
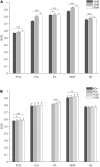
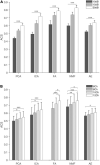
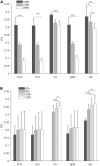
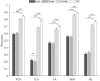
Muscle synergies have been largely used in many application fields, including motor control studies, prosthesis control, movement classification, rehabilitation, and clinical studies. Due to the complexity of the motor control system, the full repertoire of the underlying synergies has been identified only for some classes of movements and scenarios. Several extraction methods have been used to extract muscle synergies. However, some of these methods may not effectively capture the nonlinear relationship between muscles and impose constraints on input signals or extracted synergies. Moreover, other approaches such as autoencoders (AEs), an unsupervised neural network, were recently introduced to study bioinspired control and movement classification. In this study, we evaluated the performance of five methods for the extraction of spatial muscle synergy, namely, principal component analysis (PCA), independent component analysis (ICA), factor analysis (FA), nonnegative matrix factorization (NMF), and AEs using simulated data and a publicly available database. To analyze the performance of the considered extraction methods with respect to several factors, we generated a comprehensive set of simulated data (ground truth), including spatial synergies and temporal coefficients. The signal-to-noise ratio (SNR) and the number of channels (NoC) varied when generating simulated data to evaluate their effects on ground truth reconstruction. This study also tested the efficacy of each synergy extraction method when coupled with standard classification methods, including K-nearest neighbors (KNN), linear discriminant analysis (LDA), support vector machines (SVM), and Random Forest (RF). The results showed that both SNR and NoC affected the outputs of the muscle synergy analysis. Although AEs showed better performance than FA in variance accounted for and PCA in synergy vector similarity and activation coefficient similarity, NMF and ICA outperformed the other three methods. Classification tasks showed that classification algorithms were sensitive to synergy extraction methods, while KNN and RF outperformed the other two methods for all extraction methods; in general, the classification accuracy of NMF and PCA was higher. Overall, the results suggest selecting suitable methods when performing muscle synergy-related analysis.
Keywords: autoencoder (AE); factor analysis (FA); independent component analysis (ICA); muscle synergy; non-negative matrix factorization (NMF); principal component analysis (PCA).
Copyright © 2022 Zhao, Wen, Zhang, Atzori, Müller, Xie and Scano.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Muscle synergies for evaluating upper limb in clinical applications: A systematic review.Heliyon. 2023 May 11;9(5):e16202. doi: 10.1016/j.heliyon.2023.e16202. eCollection 2023 May. Heliyon. 2023. PMID: 37215841 Free PMC article. Review.
-
Evaluation of matrix factorisation approaches for muscle synergy extraction.Med Eng Phys. 2018 Jul;57:51-60. doi: 10.1016/j.medengphy.2018.04.003. Epub 2018 Apr 24. Med Eng Phys. 2018. PMID: 29703696
-
Matrix factorization algorithms for the identification of muscle synergies: evaluation on simulated and experimental data sets.J Neurophysiol. 2006 Apr;95(4):2199-212. doi: 10.1152/jn.00222.2005. Epub 2006 Jan 4. J Neurophysiol. 2006. PMID: 16394079
-
On identifying kinematic and muscle synergies: a comparison of matrix factorization methods using experimental data from the healthy population.J Neurophysiol. 2017 Jan 1;117(1):290-302. doi: 10.1152/jn.00435.2016. Epub 2016 Nov 16. J Neurophysiol. 2017. PMID: 27852733 Free PMC article.
-
Neuromuscular synergies in motor control in normal and poststroke individuals.Rev Neurosci. 2018 Aug 28;29(6):593-612. doi: 10.1515/revneuro-2017-0058. Rev Neurosci. 2018. PMID: 29397390 Review.
Cited by
-
The effect of body weight-supported Tai Chi Yunshou on upper limb motor function in stroke survivors based on neurobiomechanical analysis: a four-arm, parallel-group, assessors-blind randomized controlled trial protocol.Front Neurol. 2024 Jul 9;15:1395164. doi: 10.3389/fneur.2024.1395164. eCollection 2024. Front Neurol. 2024. PMID: 39045430 Free PMC article.
-
A methodological scoping review on EMG processing and synergy-based results in muscle synergy studies in Parkinson's disease.Front Bioeng Biotechnol. 2025 Jan 6;12:1445447. doi: 10.3389/fbioe.2024.1445447. eCollection 2024. Front Bioeng Biotechnol. 2025. PMID: 39834639 Free PMC article.
-
Muscle synergies for evaluating upper limb in clinical applications: A systematic review.Heliyon. 2023 May 11;9(5):e16202. doi: 10.1016/j.heliyon.2023.e16202. eCollection 2023 May. Heliyon. 2023. PMID: 37215841 Free PMC article. Review.
-
Evidence of synergy coordination patterns of upper-limb motor control in stroke patients with mild and moderate impairment.Front Physiol. 2023 Sep 11;14:1214995. doi: 10.3389/fphys.2023.1214995. eCollection 2023. Front Physiol. 2023. PMID: 37753453 Free PMC article.
References
-
- Abdi H., Williams L. J. (2010). Principal component analysis. WIREs Comp. Stat. 2 433–459. 10.1002/wics.101 - DOI
-
- Atzori M., Gijsberts A., Heynen S., Hager A.-G. M., Deriaz O., van der Smagt P., et al. (2012). “Building the Ninapro database: A resource for the biorobotics community,” in 2012 4th IEEE RAS & EMBS International Conference on Biomedical Robotics and Biomechatronics (BioRob), (Rome: IEEE; ), 1258–1265. 10.1109/BioRob.2012.6290287 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

