CLP1 is a Prognosis-Related Biomarker and Correlates With Immune Infiltrates in Rheumatoid Arthritis
- PMID: 35721104
- PMCID: PMC9201986
- DOI: 10.3389/fphar.2022.827215
CLP1 is a Prognosis-Related Biomarker and Correlates With Immune Infiltrates in Rheumatoid Arthritis
Abstract
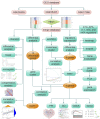
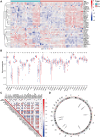
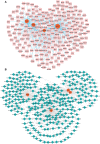
Rheumatoid arthritis (RA) is a chronic, heterogeneous autoimmune disease with a high disability rate that seriously affects society and individuals. However, there is a lack of effective and reliable diagnostic markers and therapeutic targets. In this study, we identified diagnostic markers of RA based on RNA modification and explored its role as well as degree of immune cell infiltration. We used the gene expression profile data of three synovial tissues (GSE55235, GSE55457, GSE77298) from the Gene Expression Omnibus (GEO) database and the gene of 5 RNA modification genes (including m6A, m1A, m5C, APA, A-1), combined with cluster analysis, identified four RNA modifiers closely related to RA (YTHDC1, LRPPRC, NOP2, and CLP1) and five immune cells namely T cell CD8, CD4 memory resting, T cells regulatory (Tregs) Macrophages M0, and Neutrophils. Based on the LASSO regression algorithm, hub genes and immune cell prediction models were established respectively in RA and a nomogram based on the immune cell model was built. Around 4 key RNA modification regulator genes, miRNA-mRNA, mRNA-TF networks have been established, and GSEA-GO, KEGG-GSEA enrichment analysis has been carried out. Finally, CLP1 was established as an effective RA diagnostic marker, and was highly positively correlated with T cells follicular helper (Tfh) infiltration. On the other hand, highly negatively correlated with the expression of mast cells. In short, CLP1 may play a non-negligible role in the onset and development of RA by altering immune cell infiltration, and it is predicted to represent a novel target for RA clinical diagnosis and therapy.
Keywords: CLP1; RNA modification; diagnostic biomarkers; gene expression profile data; rheumatoid arthritis.
Copyright © 2022 Zhao, He, Tang, Lai, Ren, Yu, Lin, Wang, El Akkawi, Zeng and Zha.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures











References
-
- Broeren M. G., de Vries M., Bennink M. B., Arntz O. J., Blom A. B., Koenders M. I., et al. (2016). Disease-Regulated Gene Therapy with Anti-inflammatory Interleukin-10 under the Control of the CXCL10 Promoter for the Treatment of Rheumatoid Arthritis. Hum. Gene Ther. 27 (3), 244–254. 10.1089/hum.2015.127 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

