A Toolbox for the Generation of Chemical Probes for Baculovirus IAP Repeat Containing Proteins
- PMID: 35721509
- PMCID: PMC9204419
- DOI: 10.3389/fcell.2022.886537
A Toolbox for the Generation of Chemical Probes for Baculovirus IAP Repeat Containing Proteins
Abstract
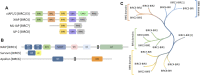
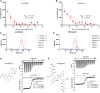
E3 ligases constitute a large and diverse family of proteins that play a central role in regulating protein homeostasis by recruiting substrate proteins via recruitment domains to the proteasomal degradation machinery. Small molecules can either inhibit, modulate or hijack E3 function. The latter class of small molecules led to the development of selective protein degraders, such as PROTACs (PROteolysis TArgeting Chimeras), that recruit protein targets to the ubiquitin system leading to a new class of pharmacologically active drugs and to new therapeutic options. Recent efforts have focused on the E3 family of Baculovirus IAP Repeat (BIR) domains that comprise a structurally conserved but diverse 70 amino acid long protein interaction domain. In the human proteome, 16 BIR domains have been identified, among them promising drug targets such as the Inhibitors of Apoptosis (IAP) family, that typically contain three BIR domains (BIR1, BIR2, and BIR3). To date, this target area lacks assay tools that would allow comprehensive evaluation of inhibitor selectivity. As a consequence, the selectivity of current BIR domain targeting inhibitors is unknown. To this end, we developed assays that allow determination of inhibitor selectivity in vitro as well as in cellulo. Using this toolbox, we have characterized available BIR domain inhibitors. The characterized chemical starting points and selectivity data will be the basis for the generation of new chemical probes for IAP proteins with well-characterized mode of action and provide the basis for future drug discovery efforts and the development of PROTACs and molecular glues.
Keywords: E3 Ligase; IAP; NanoBRET; PROTAC; Ubiquitin.
Copyright © 2022 Schwalm, Berger, Meuter, Vasta, Corona, Röhm, Berger, Farges, Beinert, Preuss, Morasch, Rogov, Mathea, Saxena, Robers, Müller and Knapp.
Conflict of interest statement
B-TB and LB are co-founders of CELLinib GmbH (Frankfurt am Main, Germany). MBR, JMW, and JDV are employees of Promega. Promega owns patents related to NanoLuc and NanoBRET Target Engagement assays. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Allensworth J. L., Sauer S. J., Lyerly H. K., Morse M. A., Devi G. R. (2013). Smac Mimetic Birinapant Induces Apoptosis and Enhances TRAIL Potency in Inflammatory Breast Cancer Cells in an IAP-dependent and TNF-α-independent Mechanism. Breast Cancer Res. Treat. 137 (2), 359–371. 10.1007/s10549-012-2352-6 - DOI - PubMed
-
- Cai Q., Sun H., Peng Y., Lu J., Nikolovska-Coleska Z., McEachern D., et al. (2011). A Potent and Orally Active Antagonist (SM-406/AT-406) of Multiple Inhibitor of Apoptosis Proteins (IAPs) in Clinical Development for Cancer Treatment. Journal of Medicinal Chemistry 54 (8), 2714–2726. 10.1016/j.febslet.2008.09.058 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

