Single Cell Transcriptomic Analysis Reveals Organ Specific Pericyte Markers and Identities
- PMID: 35722109
- PMCID: PMC9199463
- DOI: 10.3389/fcvm.2022.876591
Single Cell Transcriptomic Analysis Reveals Organ Specific Pericyte Markers and Identities
Abstract
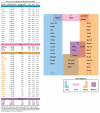
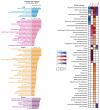
Pericytes are mesenchymal-derived mural cells that wrap around capillaries and directly contact endothelial cells. Present throughout the body, including the cardiovascular system, pericytes are proposed to have multipotent cell-like properties and are involved in numerous biological processes, including regulation of vascular development, maturation, permeability, and homeostasis. Despite their physiological importance, the functional heterogeneity, differentiation process, and pathological roles of pericytes are not yet clearly understood, in part due to the inability to reliably distinguish them from other mural cell populations. Our study focused on identifying pericyte-specific markers by analyzing single-cell RNA sequencing data from tissue-specific mouse pericyte populations generated by the Tabula Muris Senis. We identified the mural cell cluster in murine lung, heart, kidney, and bladder that expressed either of two known pericyte markers, Cspg4 or Pdgfrb. We further defined pericytes as those cells that co-expressed both markers within this cluster. Single-cell differential expression gene analysis compared this subset with other clusters that identified potential pericyte marker candidates, including Kcnk3 (in the lung); Rgs4 (in the heart); Myh11 and Kcna5 (in the kidney); Pcp4l1 (in the bladder); and Higd1b (in lung and heart). In addition, we identified novel markers of tissue-specific pericytes and signaling pathways that may be involved in maintaining their identity. Moreover, the identified markers were further validated in Human Lung Cell Atlas and human heart single-cell RNAseq databases. Intriguingly, we found that markers of heart and lung pericytes in mice were conserved in human heart and lung pericytes. In this study, we, for the first time, identified specific pericyte markers among lung, heart, kidney, and bladder and reveal differentially expressed genes and functional relationships between mural cells.
Keywords: heart; lung; pericytes; single cell RNA sequencing; tissue-specific.
Copyright © 2022 Baek, Maiorino, Kim, Glass, Raby and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Yuan K, Shao NY, Hennigs JK, Discipulo M, Orcholski ME, Shamskhou E, et al. Increased pyruvate dehydrogenase kinase 4 expression in lung pericytes is associated with reduced endothelial-pericyte interactions and small vessel loss in pulmonary arterial hypertension. Am J Pathol. (2016) 186:2500–14. 10.1016/j.ajpath.2016.05.016 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

