Comparative Genomics Provides Insights Into Genetic Diversity of Clostridium tyrobutyricum and Potential Implications for Late Blowing Defects in Cheese
- PMID: 35722315
- PMCID: PMC9201417
- DOI: 10.3389/fmicb.2022.889551
Comparative Genomics Provides Insights Into Genetic Diversity of Clostridium tyrobutyricum and Potential Implications for Late Blowing Defects in Cheese
Abstract
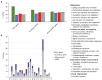
Clostridium tyrobutyricum has been recognized as the main cause of late blowing defects (LBD) in cheese leading to considerable economic losses for the dairy industry. Although differences in spoilage ability among strains of this species have been acknowledged, potential links to the genetic diversity and functional traits remain unknown. In the present study, we aimed to investigate and characterize genomic variation, pan-genomic diversity and key traits of C. tyrobutyricum by comparing the genomes of 28 strains. A comparative genomics analysis revealed an "open" pangenome comprising 9,748 genes and a core genome of 1,179 genes shared by all test strains. Among those core genes, the majority of genes encode proteins related to translation, ribosomal structure and biogenesis, energy production and conversion, and amino acid metabolism. A large part of the accessory genome is composed of sets of unique, strain-specific genes ranging from about 5 to more than 980 genes. Furthermore, functional analysis revealed several strain-specific genes related to replication, recombination and repair, cell wall, membrane and envelope biogenesis, and defense mechanisms that might facilitate survival under stressful environmental conditions. Phylogenomic analysis divided strains into two clades: clade I contained human, mud, and silage isolates, whereas clade II comprised cheese and milk isolates. Notably, these two groups of isolates showed differences in certain hypothetical proteins, transcriptional regulators and ABC transporters involved in resistance to oxidative stress. To the best of our knowledge, this is the first study to provide comparative genomics of C. tyrobutyricum strains related to LBD. Importantly, the findings presented in this study highlight the broad genetic diversity of C. tyrobutyricum, which might help us understand the diversity in spoilage potential of C. tyrobutyricum in cheese and provide some clues for further exploring the gene modules responsible for the spoilage ability of this species.
Keywords: Clostridium tyrobutyricum; cheese; comparative genomics; dairy; pangenome; spoilage.
Copyright © 2022 Podrzaj, Burtscher and Domig.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Bermudez J., Gonzalez M. J., Olivera J. A., Burgueno J. A., Juliano P., Fox E. M., et al. . (2016). Seasonal occurrence and molecular diversity of clostridia species spores along cheesemaking streams of 5 commercial dairy plants. J. Dairy Sci. 99, 3358–3366. doi: 10.3168/jds.2015-10079, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

