Identification of genetic polymorphisms in unexplained recurrent spontaneous abortion based on whole exome sequencing
- PMID: 35722368
- PMCID: PMC9201170
- DOI: 10.21037/atm-22-2179
Identification of genetic polymorphisms in unexplained recurrent spontaneous abortion based on whole exome sequencing
Abstract
Background: The precise etiology of approximately 50% of patients with recurrent spontaneous abortion (RSA) is unclear, known as unexplained recurrent spontaneous abortion (URSA). This study identified the genetic polymorphisms in patients with URSA.
Methods: Genomic DNA was extracted from 30 couples with URSA and 9 couples with normal reproductive history for whole exome sequencing. Variations in annotation, filtering, and prediction of harmfulness and pathogenicity were examined. Furthermore, predictions of the effects of changes in protein structure, Sanger validation, and functional enrichment analyses were performed. The missense mutated genes with significant changes in protein function, and genes with mutations of premature stop, splice site, frameshift, and in-frame indel were selected as candidate mutated genes related to URSA.
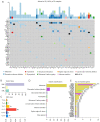
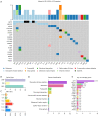
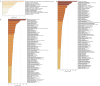
Results: In 30 unrelated couples with URSA, 50%, 20%, and 30% had 2, 3, and more than 4 miscarriages, respectively. Totally, 971 maternal and 954 paternal mutations were found to be pathogenic or possibly pathogenic after preliminary filtering. Total variations were not associated with age nor the number of miscarriages. In 28 patients (involving 23 couples), 22 pathogenic or possibly pathogenic variants of 19 genes were found to be strongly associated with URSA, with an abnormality rate of 76.67%. Among these, 12 missense variants showed obvious changes in protein functions, including ANXA5 (c.949G>C; p.G317R), APP (c.1530G>C; p.K510N), DNMT1 (c.2626G>A; p.G876R), FN1 (c.5621T>C; p.M1874T), MSH2 (c.1168G>A; p.L390F), THBS1 (c.2099A>G; p.N700S), KDR (c.2440G>A; p.D814N), POLR2B (c.406G>T; p.G136C), ITGB1 (c.655T>C; p.Y219H), PLK1 (c.1210G>T; p.A404S), COL4A2 (c.4808 A>C; p.H1603P), and LAMA4 (c.3158A>G; p.D1053G). Six other genes with mutations of premature stop, splice site, frameshift, and in-frame indel were also identified, including BUB1B (c.1648C>T; p.R550*) and MMP2 (c.1462_1464delTTC; p.F488del) from the father, and mutations from mother and/or father including BPTF (c.396_398delGGA; p.E138 del and c.429_431GGA; p.E148del), MECP2 (c.21_23delCGC; p.A7del), LAMA2 (HGVS: NA; Exon: NA; SPLICE_SITE, DONOR), and SOX21 (c.640 _641insT; p. A214fs, c.644dupC; p. A215fs and c.644_645ins ACGCGTCTTCTTCCCGCAGTC; p. A215dup).
Conclusions: These pathogenic or potentially pathogenic mutated genes may be potential biomarkers for URSA and may play an auxiliary role in the treatment of URSA.
Keywords: Whole exome sequencing; genetic polymorphisms; signaling pathway; unexplained recurrent spontaneous abortion (URSA).
2022 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2179/coif). All authors report that this study was funded by the Science and Technology Research Project of Chongqing Education Commission (No. KJQN202100427) and the Young and Middle-Aged Medical Talents Project of Chongqing Municipal Health Commission/Chongqing Science and Technology Bureau (No. 2022GDRC001). The authors have no other conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
