Transcriptomic Analysis of Long Non-coding RNA-MicroRNA-mRNA Interactions in the Nucleus Accumbens Related to Morphine Addiction in Mice
- PMID: 35722589
- PMCID: PMC9201067
- DOI: 10.3389/fpsyt.2022.915398
Transcriptomic Analysis of Long Non-coding RNA-MicroRNA-mRNA Interactions in the Nucleus Accumbens Related to Morphine Addiction in Mice
Abstract
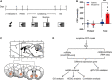
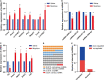
Recent research suggest that some non-coding RNAs (ncRNAs) are important regulators of chromatin dynamics and gene expression in nervous system development and neurological diseases. Nevertheless, the molecular mechanisms of long non-coding RNAs (lncRNAs), acting as competing endogenous RNAs (ceRNAs), underlying morphine addiction are still unknown. In this research, RNA sequencing (RNA-seq) was used to examine the expression profiles of lncRNAs, miRNAs and mRNAs on the nucleus accumbens (NAc) tissues of mice trained with morphine or saline conditioned place preference (CPP), with differential expression of 31 lncRNAs, 393 miRNAs, and 371 mRNAs found. A ceRNA network was established for reciprocal interactions for 9 differentially expressed lncRNAs (DElncRNAs), 10 differentially expressed miRNAs (DEmiRNAs) and 12 differentially expressed mRNAs (DEmRNAs) based on predicted miRNAs shared by lncRNAs and mRNAs. KEGG pathway enrichment analyses were conducted to explore the potential functions of DEmRNAs interacting with lncRNAs in the ceRNA network. These DEmRNAs were enriched in synaptic plasticity-related pathways, including pyrimidine metabolism, ECM-receptor interaction, and focal adhesion. The correlation between the relative expression of lncRNAs, miRNAs and mRNAs was analyzed to further validate predicted ceRNA networks, and the Lnc15qD3-miR-139-3p-Lrp2 ceRNA regulatory interaction was determined. These results suggest that the comprehensive network represents a new insight into the lncRNA-mediated ceRNA regulatory mechanisms underlying morphine addiction and provide new potential diagnostic and prognostic biomarkers for morphine addiction.
Keywords: addiction; ceRNA network; lncRNA; morphine; nucleus accumbens.
Copyright © 2022 Li, Xie, Lu, Yang, Wang, Yu, Zhang, Cong, Wen and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

