Epicardial HDAC3 Promotes Myocardial Growth Through a Novel MicroRNA Pathway
- PMID: 35722872
- PMCID: PMC9308743
- DOI: 10.1161/CIRCRESAHA.122.320785
Epicardial HDAC3 Promotes Myocardial Growth Through a Novel MicroRNA Pathway
Abstract
Background: Establishment of the myocardial wall requires proper growth cues from nonmyocardial tissues. During heart development, the epicardium and epicardium-derived cells instruct myocardial growth by secreting essential factors including FGF (fibroblast growth factor) 9 and IGF (insulin-like growth factor) 2. However, it is poorly understood how the epicardial secreted factors are regulated, in particular by chromatin modifications for myocardial formation. The current study is to investigate whether and how HDAC (histone deacetylase) 3 in the developing epicardium regulates myocardial growth.
Methods: Various cellular and mouse models in conjunction with biochemical and molecular tools were employed to study the role of HDAC3 in the developing epicardium.
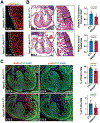
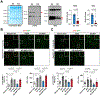
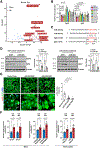
Results: We deleted Hdac3 in the developing murine epicardium, and mutant hearts showed ventricular myocardial wall hypoplasia with reduction of epicardium-derived cells. The cultured embryonic cardiomyocytes with supernatants from Hdac3 knockout (KO) mouse epicardial cells also showed decreased proliferation. Genome-wide transcriptomic analysis revealed that Fgf9 and Igf2 were significantly downregulated in Hdac3 KO mouse epicardial cells. We further found that Fgf9 and Igf2 expression is dependent on HDAC3 deacetylase activity. The supplementation of FGF9 or IGF2 can rescue the myocardial proliferation defects treated by Hdac3 KO supernatant. Mechanistically, we identified that microRNA (miR)-322 and miR-503 were upregulated in Hdac3 KO mouse epicardial cells and Hdac3 epicardial KO hearts. Overexpression of miR-322 or miR-503 repressed FGF9 and IGF2 expression, while knockdown of miR-322 or miR-503 restored FGF9 and IGF2 expression in Hdac3 KO mouse epicardial cells.
Conclusions: Our findings reveal a critical signaling pathway in which epicardial HDAC3 promotes compact myocardial growth by stimulating FGF9 and IGF2 through repressing miR-322 or miR-503, providing novel insights in elucidating the etiology of congenital heart defects and conceptual strategies to promote myocardial regeneration.
Keywords: chromatin; cues; fetal heart; pericardium; transcriptome.
Figures


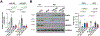

Similar articles
-
Endocardial HDAC3 is required for myocardial trabeculation.bioRxiv [Preprint]. 2024 Mar 4:2023.04.12.536668. doi: 10.1101/2023.04.12.536668. bioRxiv. 2024. Update in: Nat Commun. 2024 May 16;15(1):4166. doi: 10.1038/s41467-024-48362-6. PMID: 37886504 Free PMC article. Updated. Preprint.
-
Regenerative potential of epicardium-derived extracellular vesicles mediated by conserved miRNA transfer.Cardiovasc Res. 2022 Jan 29;118(2):597-611. doi: 10.1093/cvr/cvab054. Cardiovasc Res. 2022. PMID: 33599250 Free PMC article.
-
Retinoic acid stimulates myocardial expansion by induction of hepatic erythropoietin which activates epicardial Igf2.Development. 2011 Jan;138(1):139-48. doi: 10.1242/dev.054239. Development. 2011. PMID: 21138976 Free PMC article.
-
The epicardium in cardiac repair: from the stem cell view.Pharmacol Ther. 2011 Jan;129(1):82-96. doi: 10.1016/j.pharmthera.2010.09.002. Epub 2010 Oct 19. Pharmacol Ther. 2011. PMID: 20937304 Review.
-
The epicardium signals the way towards heart regeneration.Stem Cell Res. 2014 Nov;13(3 Pt B):683-92. doi: 10.1016/j.scr.2014.04.007. Epub 2014 Apr 29. Stem Cell Res. 2014. PMID: 24933704 Free PMC article. Review.
Cited by
-
Novel Insights into the Molecular Mechanisms Governing Embryonic Epicardium Formation.J Cardiovasc Dev Dis. 2023 Oct 24;10(11):440. doi: 10.3390/jcdd10110440. J Cardiovasc Dev Dis. 2023. PMID: 37998498 Free PMC article. Review.
-
Epigenetic determinants and non-myocardial signaling pathways contributing to heart growth and regeneration.Pharmacol Ther. 2024 May;257:108638. doi: 10.1016/j.pharmthera.2024.108638. Epub 2024 Mar 26. Pharmacol Ther. 2024. PMID: 38548089 Free PMC article. Review.
-
Specific microRNA Signature Kinetics in Porphyromonas gingivalis-Induced Periodontitis.Int J Mol Sci. 2023 Jan 24;24(3):2327. doi: 10.3390/ijms24032327. Int J Mol Sci. 2023. PMID: 36768651 Free PMC article.
-
Congenital Heart Diseases: Recent Insights into Epigenetic Mechanisms.Cells. 2025 May 31;14(11):820. doi: 10.3390/cells14110820. Cells. 2025. PMID: 40497996 Free PMC article. Review.
-
Epicardial EMT and cardiac repair: an update.Stem Cell Res Ther. 2024 Jul 19;15(1):219. doi: 10.1186/s13287-024-03823-z. Stem Cell Res Ther. 2024. PMID: 39026298 Free PMC article. Review.
References
-
- Schlueter J and Brand T. Epicardial progenitor cells in cardiac development and regeneration. J Cardiovasc Transl Res. 2012;5:641–53. - PubMed
-
- Saifi O, Ghandour B, Jaalouk D, Refaat M and Mahfouz R. Myocardial regeneration: role of epicardium and implicated genes. Mol Biol Rep. 2019;46:6661–6674. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

