Proteasomal inhibition preferentially stimulates lysosome activity relative to autophagic flux in primary astrocytes
- PMID: 35722992
- PMCID: PMC9851260
- DOI: 10.1080/15548627.2022.2084884
Proteasomal inhibition preferentially stimulates lysosome activity relative to autophagic flux in primary astrocytes
Abstract
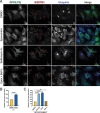
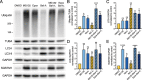
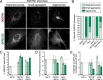
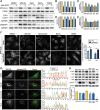
Neurons and astrocytes face unique demands on their proteome to enable proper function and survival of the nervous system. Consequently, both cell types are critically dependent on robust quality control pathways such as macroautophagy (hereafter referred to as autophagy) and the ubiquitin-proteasome system (UPS). We previously reported that autophagy is differentially regulated in astrocytes and neurons in the context of metabolic stress, but less is understood in the context of proteotoxic stress induced by inhibition of the UPS. Dysfunction of the proteasome or autophagy has been linked to the progression of various neurodegenerative diseases. Therefore, in this study, we explored the connection between autophagy and the proteasome in primary astrocytes and neurons. Prior studies largely in non-neural models report a compensatory relationship whereby inhibition of the UPS stimulates autophagy. To our surprise, inhibition of the proteasome did not robustly upregulate autophagy in astrocytes or neurons. In fact, the effects on autophagy are modest particularly in comparison to paradigms of metabolic stress. Rather, we find that UPS inhibition in astrocytes induces formation of Ub-positive aggregates that harbor the selective autophagy receptor, SQSTM1/p62, but these structures were not productive substrates for autophagy. By contrast, we observed a significant increase in lysosomal degradation in astrocytes in response to UPS inhibition, but this stimulation was not sufficient to reduce total SQSTM1 levels. Last, UPS inhibition was more toxic in neurons compared to astrocytes, suggesting a cell type-specific vulnerability to proteotoxic stress.Abbreviations: Baf A1: bafilomycin A1; CQ: chloroquine; Epox: epoxomicin; MAP1LC3/LC3: microtubule-associated protein 1 light chain 3; MTOR: mechanistic target of rapamycin kinase; p-ULK1: phospho-ULK1; SQSTM1/p62: sequestosome 1; Ub: ubiquitin; ULK1: unc-51 like kinase 1; UPS: ubiquitin-proteasome system.
Keywords: Astrocytes; LC3; SQSTM1; autophagy; lysosomes; neurons; proteasome; ubiquitin.
Conflict of interest statement
The authors declare no conflict of interest. .
Figures






References
-
- Spalding KL, Bhardwaj RD, Buchholz BA, et al. Retrospective birth dating of cells in humans. Cell. 2005. Jul 15;122(1):133–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
