N-Terminus-Mediated Solution Structure of Dimerization Domain of PRC1
- PMID: 35723369
- PMCID: PMC9164050
- DOI: 10.3390/cimb44040111
N-Terminus-Mediated Solution Structure of Dimerization Domain of PRC1
Abstract
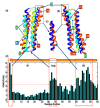
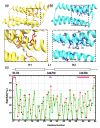
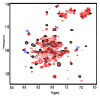
Microtubule-associated proteins (MAPs) are essential for the accurate division of a cell into two daughter cells. These proteins target specific microtubules to be incorporated into the spindle midzone, which comprises a special array of microtubules that initiate cytokinesis during anaphase. A representative member of the MAPs is Protein Regulator of Cytokinesis 1 (PRC1), which self-multimerizes to cross-link microtubules, the malfunction of which might result in cancerous cells. The importance of PRC1 multimerization makes it a popular target for structural studies. The available crystal structure of PRC1 has low resolution (>3 Å) and accuracy, limiting a better understanding of the structure-related functions of PRC1. Therefore, we used NMR spectroscopy to better determine the structure of the dimerization domain of PRC1. The NMR structure shows that the PRC1 N terminus is crucial to the overall structure integrity, but the crystal structure bespeaks otherwise. We systematically addressed the role of the N terminus by generating a series of mutants in which N-terminal residues methionine (Met1) and arginine (Arg2) were either deleted, extended or substituted with other rationally selected amino acids. Each mutant was subsequently analyzed by NMR spectroscopy or fluorescence thermal shift assays for its structural or thermal stability; we found that N-terminal perturbations indeed affected the overall protein structure and that the solution structure better reflects the conformation of PRC1 under solution conditions. These results reveal that the structure of PRC1 is governed by its N terminus through hydrophobic interactions with other core residues, such hitherto unidentified N-terminal conformations might shed light on the structure−function relationships of PRC1 or other proteins. Therefore, our study is of major importance in terms of identifying a novel structural feature and can further the progress of protein folding and protein engineering.
Keywords: N-terminal domain of PRC1; N-terminus-mediated core packing; homodimerization; hydrophobic core packing; protein regulator of cytokinesis; solution structure.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

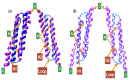



Similar articles
-
Validation of the solution structure of dimerization domain of PRC1.PLoS One. 2022 Aug 5;17(8):e0270572. doi: 10.1371/journal.pone.0270572. eCollection 2022. PLoS One. 2022. PMID: 35930764 Free PMC article.
-
Marking and measuring single microtubules by PRC1 and kinesin-4.Cell. 2013 Jul 18;154(2):377-90. doi: 10.1016/j.cell.2013.06.021. Cell. 2013. PMID: 23870126 Free PMC article.
-
PRC1 is a critical regulator for anaphase spindle midzone assembly and cytokinesis in mouse oocyte meiosis.FEBS J. 2021 May;288(9):3055-3067. doi: 10.1111/febs.15634. Epub 2020 Nov 30. FEBS J. 2021. PMID: 33206458
-
Mechanisms of the Ase1/PRC1/MAP65 family in central spindle assembly.Biol Rev Camb Philos Soc. 2019 Dec;94(6):2033-2048. doi: 10.1111/brv.12547. Epub 2019 Jul 25. Biol Rev Camb Philos Soc. 2019. PMID: 31343816 Review.
-
PRC1: Linking Cytokinesis, Chromosomal Instability, and Cancer Evolution.Trends Cancer. 2018 Jan;4(1):59-73. doi: 10.1016/j.trecan.2017.11.002. Epub 2017 Dec 16. Trends Cancer. 2018. PMID: 29413422 Review.
Cited by
-
Validation of the solution structure of dimerization domain of PRC1.PLoS One. 2022 Aug 5;17(8):e0270572. doi: 10.1371/journal.pone.0270572. eCollection 2022. PLoS One. 2022. PMID: 35930764 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

