Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair
- PMID: 35723374
- PMCID: PMC9164083
- DOI: 10.3390/cimb44040116
Validation Study to Determine the Accuracy of Widespread Promoterless EGFP Reporter at Assessing CRISPR/Cas9-Mediated Homology Directed Repair
Abstract
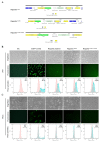
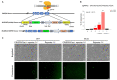
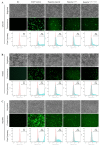
An accurate visual reporter system to assess homology-directed repair (HDR) is a key prerequisite for evaluating the efficiency of Cas9-mediated precise gene editing. Herein, we tested the utility of the widespread promoterless EGFP reporter to assess the efficiency of CRISPR/Cas9-mediated homologous recombination by fluorescence expression. We firstly established a promoterless EGFP reporter donor targeting the porcine GAPDH locus to study CRISPR/Cas9-mediated homologous recombination in porcine cells. Curiously, EGFP was expressed at unexpectedly high levels from the promoterless donor in porcine cells, with or without Cas9/sgRNA. Even higher EGFP expression was detected in human cells and those of other species when the porcine donor was transfected alone. Therefore, EGFP could be expressed at certain level in various cells transfected with the promoterless EGFP reporter alone, making it a low-resolution reporter for measuring Cas9-mediated HDR events. In summary, the widespread promoterless EGFP reporter could not be an ideal measurement for HDR screening and there is an urgent need to develop a more reliable, high-resolution HDR screening system to better explore strategies of increasing the efficiency of Cas9-mediated HDR in mammalian cells.
Keywords: CRISPR/Cas9; homology-directed repair; precise gene editing; promoterless EGFP reporter; random integration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
- 31871292/National Natural Science Foundation of China
- 2019MS089/Fundamental Research Funds for the Central Universities from South China University of Technology
- 2019A1515010619/Natural Science Foundation of Guangdong Province of China
- A2019432/Medical Scientific Research Foundation of Guangdong Province of China
- 2018YFC0910201/The National Key R&D Program of China
LinkOut - more resources
Full Text Sources
Research Materials

