Bacterial Butyrate in Parkinson's Disease Is Linked to Epigenetic Changes and Depressive Symptoms
- PMID: 35723531
- PMCID: PMC9545646
- DOI: 10.1002/mds.29128
Bacterial Butyrate in Parkinson's Disease Is Linked to Epigenetic Changes and Depressive Symptoms
Abstract
Background: The gut microbiome and its metabolites can impact brain health and are altered in Parkinson's disease (PD) patients. It has been recently demonstrated that PD patients have reduced fecal levels of the potent epigenetic modulator butyrate and its bacterial producers.
Objectives: Here, we investigate whether the changes in the gut microbiome and associated metabolites are related to PD symptoms and epigenetic markers in leucocytes and neurons.
Methods: Stool, whole blood samples, and clinical data were collected from 55 PD patients and 55 controls. We performed DNA methylation analysis on whole blood samples and analyzed the results in relation to fecal short-chain fatty acid concentrations and microbiota composition. In another cohort, prefrontal cortex neurons were isolated from control and PD brains. We identified genome-wide DNA methylation by targeted bisulfite sequencing.
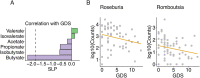
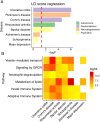
Results: We show that lower fecal butyrate and reduced counts of genera Roseburia, Romboutsia, and Prevotella are related to depressive symptoms in PD patients. Genes containing butyrate-associated methylation sites include PD risk genes and significantly overlap with sites epigenetically altered in PD blood leucocytes, predominantly neutrophils, and in brain neurons, relative to controls. Moreover, butyrate-associated methylated-DNA regions in PD overlap with those altered in gastrointestinal (GI), autoimmune, and psychiatric diseases.
Conclusions: Decreased levels of bacterially produced butyrate are related to epigenetic changes in leucocytes and neurons from PD patients and to the severity of their depressive symptoms. PD shares common butyrate-dependent epigenetic changes with certain GI and psychiatric disorders, which could be relevant for their epidemiological relation. © 2022 The Authors. Movement Disorders published by Wiley Periodicals LLC on behalf of International Parkinson and Movement Disorder Society.
Keywords: DNA methylation; Parkinson's disease; epigenetics; gut brain axis; microbiome.
© 2022 The Authors. Movement Disorders published by Wiley Periodicals LLC on behalf of International Parkinson and Movement Disorder Society.
Figures



Comment in
-
Epigenetic Drug Effects in Levodopa-Treated Patients with Parkinson's Disease.Mov Disord. 2023 Apr;38(4):710-711. doi: 10.1002/mds.29365. Mov Disord. 2023. PMID: 37061885 No abstract available.
References
-
- Svensson E, Horváth‐Puhó E, Thomsen RW, et al. Vagotomy and subsequent risk of Parkinson's disease. Ann Neurol 2015;78:522–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

