Rational domestication of a plant-based recombinant expression system expands its biosynthetic range
- PMID: 35724659
- PMCID: PMC9578353
- DOI: 10.1093/jxb/erac273
Rational domestication of a plant-based recombinant expression system expands its biosynthetic range
Abstract
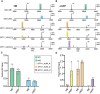
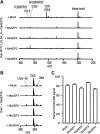
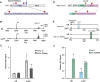
Plant molecular farming aims to provide a green, flexible, and rapid alternative to conventional recombinant expression systems, capable of producing complex biologics such as enzymes, vaccines, and antibodies. Historically, the recombinant expression of therapeutic peptides in plants has proven difficult, largely due to their small size and instability. However, some plant species harbour the capacity for peptide backbone cyclization, a feature inherent in stable therapeutic peptides. One obstacle to realizing the potential of plant-based therapeutic peptide production is the proteolysis of the precursor before it is matured into its final stabilized form. Here we demonstrate the rational domestication of Nicotiana benthamiana within two generations to endow this plant molecular farming host with an expanded repertoire of peptide sequence space. The in planta production of molecules including an insecticidal peptide, a prostate cancer therapeutic lead, and an orally active analgesic is demonstrated.
Keywords: Asparaginyl endopeptidase (AEP); CRISPR/Cas9; cyclotide; gene editing; insecticide; peptide; plant molecular farming; protease; recombinant; therapeutic.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
-
- Bally J, Jung H, Mortimer C, Naim F, Philips JG, Hellens R, Bombarely A, Goodin MM, Waterhouse PM.. 2018. The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annual Review of Phytopathology 56, 405–426. - PubMed
-
- Barone PW, Wiebe ME, Leung JC, et al. . 2020. Viral contamination in biologic manufacture and implications for emerging therapies. Nature Biotechnology 38, 563–572. - PubMed
-
- Bray NL, Pimentel H, Melsted P, Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology 34, 525–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

