On the caveats of a multiplex test for SARS-CoV-2 to detect seroconversion after infection or vaccination
- PMID: 35725758
- PMCID: PMC9208546
- DOI: 10.1038/s41598-022-14294-8
On the caveats of a multiplex test for SARS-CoV-2 to detect seroconversion after infection or vaccination
Abstract
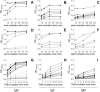
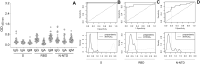
The Covid-19 pandemic, caused by SARS-CoV-2, has resulted in over 6 million reported deaths worldwide being one of the biggest challenges the world faces today. Here we present optimizations of all steps of an enzyme-linked immunosorbent assay (ELISA)-based test to detect IgG, IgA and IgM against the trimeric spike (S) protein, receptor binding domain (RBD), and N terminal domain of the nucleocapsid (N-NTD) protein of SARS-CoV-2. We discuss how to determine specific thresholds for antibody positivity and its limitations according to the antigen used. We applied the assay to a cohort of 126 individuals from Rio de Janeiro, Brazil, consisting of 23 PCR-positive individuals and 103 individuals without a confirmed diagnosis for SARS-CoV-2 infection. To illustrate the differences in serological responses to vaccinal immunization, we applied the test in 18 individuals from our cohort before and after receiving ChAdOx-1 nCoV-19 or CoronaVac vaccines. Taken together, our results show that the test can be customized at different stages depending on its application, enabling the user to analyze different cohorts, saving time, reagents, or samples. It is also a valuable tool for elucidating the immunological consequences of new viral strains and monitoring vaccination coverage and duration of response to different immunization regimens.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO. WHO Coronavirus (COVID-19) Dashboard.https://covid19.who.int/
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

