M-CSF as a therapeutic target in BRAFV600E melanoma resistant to BRAF inhibitors
- PMID: 35725813
- PMCID: PMC9470708
- DOI: 10.1038/s41416-022-01886-4
M-CSF as a therapeutic target in BRAFV600E melanoma resistant to BRAF inhibitors
Abstract
Background: Disseminated BRAFV600E melanoma responds to BRAF inhibitors (BRAFi) but easily develops resistance with poor prognosis. Secretome plays a pivotal role during tumour progression causing profound effects on therapeutic efficacy. Secreted M-CSF is involved in both cytotoxicity suppression and tumour progression in melanoma. We aimed to analyse the M-CSF contribution in resistant metastatic melanoma to BRAF-targeted therapies.
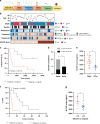
Methods: Conditioned media from melanoma cells were analysed by citoarray. Viability and migration/invasion assays were performed with paired melanoma cells and tumour growth in xenografted SCID mice. We evaluated the impact of M-CSF plasma levels with clinical prognosis from 35 metastatic BRAFV600E-mutant melanoma patients.
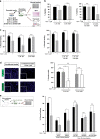
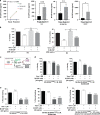
Results: BRAFi-resistant melanoma cells secretome is rich in pro-tumour cytokines. M-CSF secretion is essential to induce a Vemurafenib-resistant phenotype in melanoma cells. Further, we demonstrated that M-CSF mAb in combination with Vemurafenib and autophagy blockers synergistically induce apoptosis, impair migration and reduce tumour growth in BRAFi-resistant melanoma cells. Interestingly, lower M-CSF plasma levels are associated with better prognosis in metastatic melanoma patients.
Conclusions: Secreted M-CSF induces a BRAFi-resistant phenotype and means worse prognosis in BRAFV600E metastatic melanoma patients. These results identify secreted M-CSF as a promising therapeutic target toward BRAFi-resistant melanomas.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Kakadia S, Yarlagadda N, Awad R, Kundranda M, Niu J, Naraev B, et al. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of us food and drug administration-approved targeted therapy in advanced melanoma. Onco Targets Ther. 2018;11:7095–107. doi: 10.2147/OTT.S182721. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

