Cyanophages from a less virulent clade dominate over their sister clade in global oceans
- PMID: 35726021
- PMCID: PMC9381782
- DOI: 10.1038/s41396-022-01259-y
Cyanophages from a less virulent clade dominate over their sister clade in global oceans
Abstract
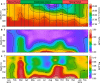
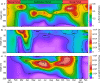
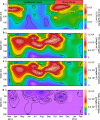
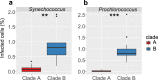
Environmental virus communities are highly diverse. However, the infection physiology underlying the evolution of diverse phage lineages and their ecological consequences are largely unknown. T7-like cyanophages are abundant in nature and infect the marine unicellular cyanobacteria, Synechococcus and Prochlorococcus, important primary producers in the oceans. Viruses belonging to this genus are divided into two distinct phylogenetic clades: clade A and clade B. These viruses have narrow host-ranges with clade A phages primarily infecting Synechococcus genotypes, while clade B phages are more diverse and can infect either Synechococcus or Prochlorococcus genotypes. Here we investigated infection properties (life history traits) and environmental abundances of these two clades of T7-like cyanophages. We show that clade A cyanophages have more rapid infection dynamics, larger burst sizes and greater virulence than clade B cyanophages. However, clade B cyanophages were at least 10-fold more abundant in all seasons, and infected more cyanobacteria, than clade A cyanophages in the Red Sea. Models predicted that steady-state cyanophage abundances, infection frequency, and virus-induced mortality, peak at intermediate virulence values. Our findings indicate that differences in infection properties are reflected in virus phylogeny at the clade level. They further indicate that infection properties, together with differences in subclade diversity and host repertoire, have important ecological consequences with the less aggressive, more diverse virus clade having greater ecological impacts.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Aiewsakun P, Adriaenssens EM, Lavigne R, Kropinski AM, Simmonds P. Evaluation of the genomic diversity of viruses infecting bacteria, archaea and eukaryotes using a common bioinformatic platform: Steps towards a unified taxonomy. J Gen Virol. 2018;99:1331–43. doi: 10.1099/jgv.0.001110. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- ERC-CoG 646868/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- 749/11/Israel Science Foundation (ISF)
- 2679/20/Israel Science Foundation (ISF)
- 721254/Simons Foundation
- 721231/Simons Foundation

