A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth
- PMID: 35726026
- PMCID: PMC9236904
- DOI: 10.1038/s42255-022-00584-y
A YAP/TAZ-TEAD signalling module links endothelial nutrient acquisition to angiogenic growth
Abstract
Angiogenesis, the process by which endothelial cells (ECs) form new blood vessels from existing ones, is intimately linked to the tissue's metabolic milieu and often occurs at nutrient-deficient sites. However, ECs rely on sufficient metabolic resources to support growth and proliferation. How endothelial nutrient acquisition and usage are regulated is unknown. Here we show that these processes are instructed by Yes-associated protein 1 (YAP)/WW domain-containing transcription regulator 1 (WWTR1/TAZ)-transcriptional enhanced associate domain (TEAD): a transcriptional module whose function is highly responsive to changes in the tissue environment. ECs lacking YAP/TAZ or their transcriptional partners, TEAD1, 2 and 4 fail to divide, resulting in stunted vascular growth in mice. Conversely, activation of TAZ, the more abundant paralogue in ECs, boosts proliferation, leading to vascular hyperplasia. We find that YAP/TAZ promote angiogenesis by fuelling nutrient-dependent mTORC1 signalling. By orchestrating the transcription of a repertoire of cell-surface transporters, including the large neutral amino acid transporter SLC7A5, YAP/TAZ-TEAD stimulate the import of amino acids and other essential nutrients, thereby enabling mTORC1 activation. Dissociating mTORC1 from these nutrient inputs-elicited by the loss of Rag GTPases-inhibits mTORC1 activity and prevents YAP/TAZ-dependent vascular growth. Together, these findings define a pivotal role for YAP/TAZ-TEAD in controlling endothelial mTORC1 and illustrate the essentiality of coordinated nutrient fluxes in the vasculature.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
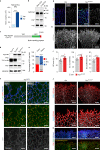
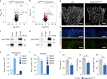


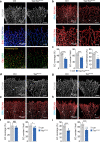
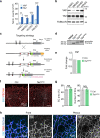
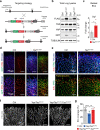

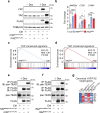
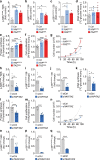

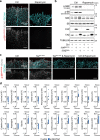

Comment in
-
YAP/TAZ-TEAD link angiogenesis to nutrients.Nat Metab. 2022 Jun;4(6):645-646. doi: 10.1038/s42255-022-00579-9. Nat Metab. 2022. PMID: 35726025 No abstract available.
References
-
- Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. 2017;357:eaal2379. - PubMed
-
- Potente M, Makinen T. Vascular heterogeneity and specialization in development and disease. Nat. Rev. Mol. Cell Biol. 2017;18:477–494. - PubMed
-
- Li X, Sun X, Carmeliet P. Hallmarks of endothelial cell metabolism in health and disease. Cell Metab. 2019;30:414–433. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

