SPIKES: Identification of physicochemical properties of spike proteins across diverse host species of SARS-CoV-2
- PMID: 35726315
- PMCID: PMC9127179
- DOI: 10.1016/j.xpro.2022.101460
SPIKES: Identification of physicochemical properties of spike proteins across diverse host species of SARS-CoV-2
Abstract
We describe a protocol to identify physicochemical properties using amino acid sequences of spike (S) proteins of SARS-CoV-2. We present an S protein prediction technique named SPIKES, incorporating an inheritable bi-objective combinatorial genetic algorithm to determine the host species specificity. This protocol addresses the S protein amino acid sequence data collection, preprocessing, methodology, and analysis. For complete details on the use and execution of this protocol, please refer to Yerukala Sathipati et al. (2022).
Keywords: Bioinformatics; Microbiology; Proteomics; Systems biology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare competing interests.
Figures
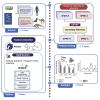






References
-
- Chang C.-C., Lin C.-J. LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011;2:1–27. doi: 10.1145/1961189.1961199. - DOI
-
- Geisow M.J., Roberts R.D.B. Amino acid preferences for secondary structure vary with protein class. Int. J. Biol. Macromol. 1980;2:387–389. doi: 10.1016/0141-8130(80)90023-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

