Ecology of Methanonatronarchaeia
- PMID: 35726892
- PMCID: PMC9796771
- DOI: 10.1111/1462-2920.16108
Ecology of Methanonatronarchaeia
Abstract
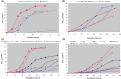
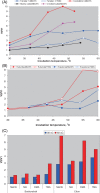
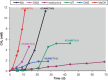
Methanonatronarchaeia represents a deep-branching phylogenetic lineage of extremely halo(alkali)philic and moderately thermophilic methyl-reducing methanogens belonging to the phylum Halobacteriota. It includes two genera, the alkaliphilic Methanonatronarchaeum and the neutrophilic Ca. Methanohalarchaeum. The former is represented by multiple closely related pure culture isolates from hypersaline soda lakes, while the knowledge about the latter is limited to a few mixed cultures with anaerobic haloarchaea. To get more insight into the distribution and ecophysiology of this enigmatic group of extremophilic methanogens, potential activity tests and enrichment cultivation with different substrates and at different conditions were performed with anaerobic sediment slurries from various hypersaline lakes in Russia. Methanonatronarchaeum proliferated exclusively in hypersaline soda lake samples mostly at elevated temperature, while at mesophilic conditions it coexisted with the extremely salt-tolerant methylotroph Methanosalsum natronophilum. Methanonatronarchaeum was also able to serve as a methylotrophic or hydrogenotrophic partner in several thermophilic enrichment cultures with fermentative bacteria. Ca. Methanohalarchaeum did not proliferate at mesophilic conditions and at thermophilic conditions it competed with extremely halophilic and moderately thermophilic methylotroph Methanohalobium, which it outcompeted at a combination of elevated temperature and methyl-reducing conditions. Overall, the results demonstrated that Methanonatronarchaeia are specialized extremophiles specifically proliferating in conditions of elevated temperature coupled with extreme salinity and simultaneous availability of a wide range of C1 -methylated compounds and H2 /formate.
© 2022 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Anisimova, M. & Gascuel, O. (2006) Approximate likelihood‐ratio test for branches: a fast, accurate, and powerful alternative. Systematic Biology, 55, 539–552. - PubMed
-
- Borrel, G. , Parisot, N. , Harris, H.M.B. , Peyretaillade, E. , Gaci, N. , Tottey, W. et al. (2015) Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales‐related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics, 15, 679. - PMC - PubMed
-
- Buchfink, B. , Xie, C. & Huson, D.H. (2015) Fast and sensitive protein alignment using DIAMOND. Nature Methods, 12, 59–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

