Balancing nutrient and energy demand and supply via autophagy
- PMID: 35728554
- PMCID: PMC9652773
- DOI: 10.1016/j.cub.2022.04.071
Balancing nutrient and energy demand and supply via autophagy
Abstract
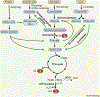
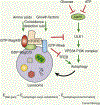
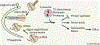
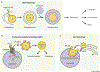
Maintaining nutrient and energy homeostasis is crucial for the survival and function of cells and organisms in response to environmental stress. Cells have evolved a stress-induced catabolic pathway, termed autophagy, to adapt to stress conditions such as starvation. During autophagy, damaged or non-essential cellular structures are broken down in lysosomes, and the resulting metabolites are reused for core biosynthetic processes or energy production. Recent studies have revealed that autophagy can target and degrade different types of nutrient stores and produce a variety of metabolites and fuels, including amino acids, nucleotides, lipids and carbohydrates. Here, we will focus on how autophagy functions to balance cellular nutrient and energy demand and supply - specifically, how energy deprivation switches on autophagic catabolism, how autophagy halts anabolism by degrading the protein synthesis machinery, and how bulk and selective autophagy-derived metabolites recycle and feed into a variety of bioenergetic and anabolic pathways during stress conditions. Recent new insights and progress in these areas provide a better understanding of how resource mobilization and reallocation sustain essential metabolic and anabolic activities under unfavorable conditions.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The author declares no competing interests.
Figures
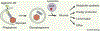
References
-
- Kuma A, and Mizushima N (2010). Physiological role of autophagy as an intracellular recycling system: With an emphasis on nutrient metabolism. Semin Cell Dev Biol 21, 683–690. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

