J-domain protein chaperone circuits in proteostasis and disease
- PMID: 35729039
- PMCID: PMC9759622
- DOI: 10.1016/j.tcb.2022.05.004
J-domain protein chaperone circuits in proteostasis and disease
Abstract
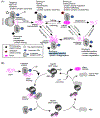
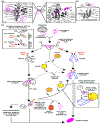
The J-domain proteins (JDP) form the largest protein family among cellular chaperones. In cooperation with the Hsp70 chaperone system, these co-chaperones orchestrate a plethora of distinct functions, including those that help maintain cellular proteostasis and development. JDPs evolved largely through the fusion of a J-domain with other protein subdomains. The highly conserved J-domain facilitates the binding and activation of Hsp70s. How JDPs (re)wire Hsp70 chaperone circuits and promote functional diversity remains insufficiently explained. Here, we discuss recent advances in our understanding of the JDP family with a focus on the regulation built around J-domains to ensure correct pairing and assembly of JDP-Hsp70 machineries that operate on different clientele under various cellular growth conditions.
Keywords: Hsp40; Hsp70; J-domain; J-domain proteins; protein conformational diseases; protein homeostasis.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

