Alu RNA induces NLRP3 expression through TLR7 activation in α-1-antitrypsin-deficient macrophages
- PMID: 35730566
- PMCID: PMC9309046
- DOI: 10.1172/jci.insight.158791
Alu RNA induces NLRP3 expression through TLR7 activation in α-1-antitrypsin-deficient macrophages
Abstract
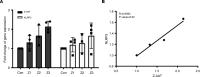
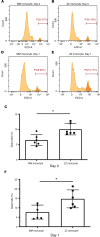
α-1 antitrypsin (AAT) is a serine protease inhibitor that plays a pivotal role in maintaining lung homeostasis. The most common AAT allele associated with AAT deficiency (AATD) is PiZ. Z-AAT accumulates in cells due to misfolding, causing severe AATD. The major function of AAT is to neutralize neutrophil elastase in the lung. It is generally accepted that loss of antiprotease function is a major cause of COPD in individuals with AATD. However, it is now being recognized that the toxic gain-of-function effect of Z-AAT in macrophage likely contributes to lung disease. In the present study, we determined that TLR7 signaling is activated in Z-MDMs, and the expression level of NLRP3, one of the targets of TLR7 signaling, is significantly higher in Z- compared with M-MDMs. We also determined that the level of endosomal Alu RNA is significantly higher in Z-compared with M-MDMs. Alu RNA is a known endogenous ligand that activates TLR7 signaling. Z-AAT likely induces the expression of Alu elements in MDMs and accelerates monocyte death, leading to the higher level of endosomal Alu RNA in Z-MDMs. Taken together,this study identifies a mechanism responsible for the toxic gain of function of Z-AAT macrophages.
Keywords: Cell Biology; Inflammation; Macrophages.
Conflict of interest statement
Figures






Similar articles
-
Alpha 1 Antitrypsin-Deficient Macrophages Have Impaired Efferocytosis of Apoptotic Neutrophils.Front Immunol. 2020 Nov 20;11:574410. doi: 10.3389/fimmu.2020.574410. eCollection 2020. Front Immunol. 2020. PMID: 33329539 Free PMC article.
-
Alpha-1 Antitrypsin-Deficient Macrophages Have Increased Matriptase-Mediated Proteolytic Activity.Am J Respir Cell Mol Biol. 2017 Aug;57(2):238-247. doi: 10.1165/rcmb.2016-0366OC. Am J Respir Cell Mol Biol. 2017. PMID: 28362108 Free PMC article.
-
Alpha-defensins inhibit ERK/STAT3 signaling during monocyte-macrophage differentiation and impede macrophage function.Respir Res. 2023 Dec 11;24(1):309. doi: 10.1186/s12931-023-02605-0. Respir Res. 2023. PMID: 38082274 Free PMC article.
-
Alpha-1-antitrypsin deficiency: Genetic variations, clinical manifestations and therapeutic interventions.Mutat Res Rev Mutat Res. 2017 Jul;773:14-25. doi: 10.1016/j.mrrev.2017.03.001. Epub 2017 Mar 18. Mutat Res Rev Mutat Res. 2017. PMID: 28927525 Review.
-
Protease-Specific Biomarkers to Analyse Protease Inhibitors for Emphysema Associated with Alpha 1-Antitrypsin Deficiency. An Overview of Current Approaches.Int J Mol Sci. 2021 Jan 21;22(3):1065. doi: 10.3390/ijms22031065. Int J Mol Sci. 2021. PMID: 33494436 Free PMC article. Review.
Cited by
-
TLR7 promotes smoke-induced experimental lung damage through the activity of mast cell tryptase.Nat Commun. 2023 Nov 14;14(1):7349. doi: 10.1038/s41467-023-42913-z. Nat Commun. 2023. PMID: 37963864 Free PMC article.
-
Ro60-Roles in RNA Processing, Inflammation, and Rheumatic Autoimmune Diseases.Int J Mol Sci. 2024 Jul 14;25(14):7705. doi: 10.3390/ijms25147705. Int J Mol Sci. 2024. PMID: 39062948 Free PMC article. Review.
-
Type 1 diabetes contributes to combined pulmonary fibrosis and emphysema in male alpha 1 antitrypsin deficient mice.PLoS One. 2023 Oct 11;18(10):e0291948. doi: 10.1371/journal.pone.0291948. eCollection 2023. PLoS One. 2023. PMID: 37819895 Free PMC article.
-
NAFLD and AATD Are Two Diseases with Unbalanced Lipid Metabolism: Similarities and Differences.Biomedicines. 2023 Jul 12;11(7):1961. doi: 10.3390/biomedicines11071961. Biomedicines. 2023. PMID: 37509601 Free PMC article. Review.
-
Monocyte NLRP3 inflammasome and interleukin-1β activation modulated by alpha-1 antitrypsin therapy in deficient individuals.Thorax. 2024 Aug 19;79(9):822-833. doi: 10.1136/thorax-2023-221071. Thorax. 2024. PMID: 38418195 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

