One Health Genomic Analysis of Extended-Spectrum β-Lactamase‒Producing Salmonella enterica, Canada, 2012‒2016
- PMID: 35731173
- PMCID: PMC9239887
- DOI: 10.3201/eid2807.211528
One Health Genomic Analysis of Extended-Spectrum β-Lactamase‒Producing Salmonella enterica, Canada, 2012‒2016
Abstract
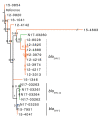
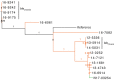
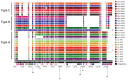
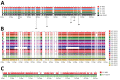
Extended-spectrum β-lactamases (ESBLs) confer resistance to extended-spectrum cephalosporins, a major class of clinical antimicrobial drugs. We used genomic analysis to investigate whether domestic food animals, retail meat, and pets were reservoirs of ESBL-producing Salmonella for human infection in Canada. Of 30,303 Salmonella isolates tested during 2012-2016, we detected 95 ESBL producers. ESBL serotypes and alleles were mostly different between humans (n = 54) and animals/meat (n = 41). Two exceptions were blaSHV-2 and blaCTX-M-1 IncI1 plasmids, which were found in both sources. A subclade of S. enterica serovar Heidelberg isolates carrying the same IncI1-blaSHV-2 plasmid differed by only 1-7 single nucleotide variants. The most common ESBL producer in humans was Salmonella Infantis carrying blaCTX-M-65, which has since emerged in poultry in other countries. There were few instances of similar isolates and plasmids, suggesting that domestic animals and retail meat might have been minor reservoirs of ESBL-producing Salmonella for human infection.
Keywords: Canada; ESBL; One Health; Salmonella enterica; antimicrobial resistance; bacteria; extended-spectrum β-lactamases; food animals; food safety; food-borne infections; genomic analysis; pets; retail meats; zoonoses.
Figures

References
-
- Roth GA, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, et al.; GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1736–88. 10.1016/S0140-6736(18)32203-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

