Functional characterization of co-phosphorylation networks
- PMID: 35731218
- PMCID: PMC9344848
- DOI: 10.1093/bioinformatics/btac406
Functional characterization of co-phosphorylation networks
Abstract
Motivation: Protein phosphorylation is a ubiquitous regulatory mechanism that plays a central role in cellular signaling. According to recent estimates, up to 70% of human proteins can be phosphorylated. Therefore, the characterization of phosphorylation dynamics is critical for understanding a broad range of biological and biochemical processes. Technologies based on mass spectrometry are rapidly advancing to meet the needs for high-throughput screening of phosphorylation. These technologies enable untargeted quantification of thousands of phosphorylation sites in a given sample. Many labs are already utilizing these technologies to comprehensively characterize signaling landscapes by examining perturbations with drugs and knockdown approaches, or by assessing diverse phenotypes in cancers, neuro-degerenational diseases, infectious diseases and normal development.
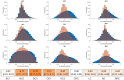
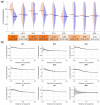
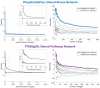
Results: We comprehensively investigate the concept of 'co-phosphorylation' (Co-P), defined as the correlated phosphorylation of a pair of phosphosites across various biological states. We integrate nine publicly available phosphoproteomics datasets for various diseases (including breast cancer, ovarian cancer and Alzheimer's disease) and utilize functional data related to sequence, evolutionary histories, kinase annotations and pathway annotations to investigate the functional relevance of Co-P. Our results across a broad range of studies consistently show that functionally associated sites tend to exhibit significant positive or negative Co-P. Specifically, we show that Co-P can be used to predict with high precision the sites that are on the same pathway or that are targeted by the same kinase. Overall, these results establish Co-P as a useful resource for analyzing phosphoproteins in a network context, which can help extend our knowledge on cellular signaling and its dysregulation.
Availability and implementation: github.com/msayati/Cophosphorylation. This research used the publicly available datasets published by other researchers as cited in the manuscript.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2022. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures