Auto-encoded Latent Representations of White Matter Streamlines for Quantitative Distance Analysis
- PMID: 35731372
- PMCID: PMC9588484
- DOI: 10.1007/s12021-022-09593-4
Auto-encoded Latent Representations of White Matter Streamlines for Quantitative Distance Analysis
Abstract
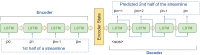
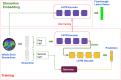
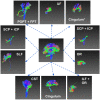
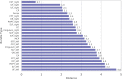
Parcellation of whole brain tractograms is a critical step to study brain white matter structures and connectivity patterns. The existing methods based on supervised classification of streamlines into predefined streamline bundle types are not designed to explore sub-bundle structures, and methods with manually designed features are expensive to compute streamline-wise similarities. To resolve these issues, we propose a novel atlas-free method that learns a latent space using a deep recurrent auto-encoder trained in an unsupervised manner. The method efficiently embeds any length of streamlines to fixed-size feature vectors, named streamline embedding, for tractogram parcellation using non-parametric clustering in the latent space. The method was evaluated on the ISMRM 2015 tractography challenge dataset with discrimination of major bundles using clustering algorithms and streamline querying based on similarity, as well as real tractograms of 102 subjects Human Connectome Project. The learnt latent streamline and bundle representations open the possibility of quantitative studies of arbitrary granularity of sub-bundle structures using generic data mining techniques.
Keywords: Diffusion MRI; Recurrent auto-encoder; Streamline clustering; Streamline embedding; Streamline tractography.
© 2022. The Author(s).
Conflict of interest statement
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose.
Figures







References
-
- Alexandroni, G., Podolsky, Y., Greenspan, H., Remez, T., Litany, O., Bronstein, A., & Giryes, R. (2017, September). White matter fiber representation using continuous dictionary learning. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 566–574). Springer, Cham.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

