Single-Cell Chemical Proteomics (SCCP) Interrogates the Timing and Heterogeneity of Cancer Cell Commitment to Death
- PMID: 35731985
- PMCID: PMC9260713
- DOI: 10.1021/acs.analchem.2c00413
Single-Cell Chemical Proteomics (SCCP) Interrogates the Timing and Heterogeneity of Cancer Cell Commitment to Death
Abstract
Chemical proteomics studies the effects of drugs upon a cellular proteome. Due to the complexity and diversity of tumors, the response of cancer cells to drugs is also heterogeneous, and thus, proteome analysis at the single-cell level is needed. Here, we demonstrate that single-cell proteomics techniques have become quantitative enough to tackle the drug effects on target proteins, enabling single-cell chemical proteomics (SCCP). Using SCCP, we studied here the time-resolved response of individual adenocarcinoma A549 cells to anticancer drugs methotrexate, camptothecin, and tomudex, revealing the early emergence of cellular subpopulations committed and uncommitted to death. As a novel and useful approach to exploring the heterogeneous response to drugs of cancer cells, SCCP may prove to be a breakthrough application for single-cell proteomics.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
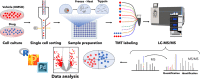
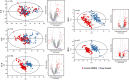
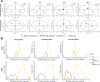


References
-
- Saei A. A.; Sabatier P.; Tokat Ü. G.; Chernobrovkin A.; Pirmoradian M.; Zubarev R. A. Comparative Proteomics of Dying and Surviving Cancer Cells Improves the Identification of Drug Targets and Sheds Light on Cell Life/Death Decisions. Mol. Cell. Proteomics 2018, 17, 1144–1155. 10.1074/mcp.RA118.000610. - DOI - PMC - PubMed
-
- Saei A. A.; Beusch C. M.; Chernobrovkin A.; Sabatier P.; Zhang B.; Tokat Ü. G.; Stergiou E.; Gaetani M.; Végvári Á.; Zubarev R. A. ProTargetMiner as a Proteome Signature Library of Anticancer Molecules for Functional Discovery. Nat. Commun. 2019, 10, 5715 10.1038/s41467-019-13582-8. - DOI - PMC - PubMed
-
- Gaetani M.; Zubarev R.. New Promises of Chemical Proteomics for Drug Development. In Novel Approaches in Drug Designing & Development, Juniper Publishers Inc., 2017; Vol. 2, pp 12–14.
-
- Ye C.; Ho D. J.; Neri M.; Yang C.; Kulkarni T.; Randhawa R.; Henault M.; Mostacci N.; Farmer P.; Renner S.; Ihry R.; Mansur L.; Keller C. G.; McAllister G.; Hild M.; Jenkins J.; Kaykas A. DRUG-Seq for Miniaturized High-Throughput Transcriptome Profiling in Drug Discovery. Nat. Commun. 2018, 9, 4307 10.1038/s41467-018-06500-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

