Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike
- PMID: 35732171
- PMCID: PMC9174147
- DOI: 10.1016/j.celrep.2022.111009
Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike
Abstract
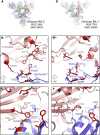
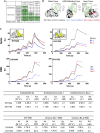
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.2 sub-lineage has gained in proportion relative to BA.1. Because spike (S) protein variations may underlie differences in their pathobiology, here we determine cryoelectron microscopy (cryo-EM) structures of the BA.2 S ectodomain and compare these with previously determined BA.1 S structures. BA.2 receptor-binding domain (RBD) mutations induce remodeling of the RBD structure, resulting in tighter packing and improved thermostability. Interprotomer RBD interactions are enhanced in the closed (or 3-RBD-down) BA.2 S, while the fusion peptide is less accessible to antibodies than in BA.1. Binding and pseudovirus neutralization assays reveal extensive immune evasion while defining epitopes of two outer RBD face-binding antibodies, DH1044 and DH1193, that neutralize both BA.1 and BA.2. Taken together, our results indicate that stabilization of the closed state through interprotomer RBD-RBD packing is a hallmark of the Omicron variant and show differences in key functional regions in the BA.1 and BA.2 S proteins.
Keywords: CP: Microbiology; Omicron BA.2; SARS-CoV-2 spike; cryoelectron microscopy; fusion peptide; immune evasion; receptor binding domain.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.F.H., K.O.S., B.K., X.L., R.J.E., S.M.-C.G., and P.A. are named in patents submitted on the SARS-CoV-2 monoclonal antibodies studied in this paper. Other authors declare no competing interests.
Figures






References
-
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. - DOI - PMC - PubMed
-
- Case J.B., Mackin S., Errico J., Chong Z., Madden E.A., Guarino B., Schmid M.A., Rosenthal K., Ren K., Jung A., et al. Resilience of S309 and AZD7442 monoclonal antibody treatments against infection by SARS-CoV-2 Omicron lineage strains. bioRxiv. 2022 doi: 10.1101/2022.03.17.484787. Preprint at. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

