Supramolecular assembly of GSK3α as a cellular response to amino acid starvation
- PMID: 35732190
- PMCID: PMC9357031
- DOI: 10.1016/j.molcel.2022.05.025
Supramolecular assembly of GSK3α as a cellular response to amino acid starvation
Abstract
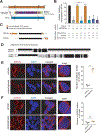
The tolerance of amino acid starvation is fundamental to robust cellular fitness. Asparagine depletion is lethal to some cancer cells, a vulnerability that can be exploited clinically. We report that resistance to asparagine starvation is uniquely dependent on an N-terminal low-complexity domain of GSK3α, which its paralog GSK3β lacks. In response to depletion of specific amino acids, including asparagine, leucine, and valine, this domain mediates supramolecular assembly of GSK3α with ubiquitin-proteasome system components in spatially sequestered cytoplasmic bodies. This effect is independent of mTORC1 or GCN2. In normal cells, GSK3α promotes survival during essential amino acid starvation. In human leukemia, GSK3α body formation predicts asparaginase resistance, and sensitivity to asparaginase combined with a GSK3α inhibitor. We propose that GSK3α body formation provides a cellular mechanism to maximize the catalytic efficiency of proteasomal protein degradation in response to amino acid starvation, an adaptive response co-opted by cancer cells for asparaginase resistance.
Keywords: GSK3; Wnt; asparaginase; protein degradation; ubiquitin-proteasome system.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Boston Children’s Hospital has filed patents on the subject matter of this manuscript. G.C. is a founder of Samus Therapeutics and a member of its board of directors. A.G. is on a scientific advisory board for Attivare Therapeutics and receives research funding from Astellas Pharma.
Figures





References
-
- Acebron SP, Karaulanov E, Berger BS, Huang YL, and Niehrs C (2014). Mitotic wnt signaling promotes protein stabilization and regulates cell size. Mol Cell 54, 663–674. - PubMed
-
- Ahmad F, and Woodgett JR (2020). Emerging roles of GSK-3alpha in pathophysiology: Emphasis on cardio-metabolic disorders. Biochim Biophys Acta Mol Cell Res 1867, 118616. - PubMed
-
- Appel IM, den Boer ML, Meijerink JP, Veerman AJ, Reniers NC, and Pieters R (2006). Up-regulation of asparagine synthetase expression is not linked to the clinical response L-asparaginase in pediatric acute lymphoblastic leukemia. Blood 107, 4244–4249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

