N6 -methyladenosine-modified circRNA RERE modulates osteoarthritis by regulating β-catenin ubiquitination and degradation
- PMID: 35733354
- PMCID: PMC9816929
- DOI: 10.1111/cpr.13297
N6 -methyladenosine-modified circRNA RERE modulates osteoarthritis by regulating β-catenin ubiquitination and degradation
Abstract
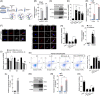
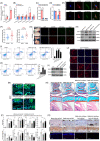
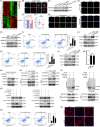
Objectives: N6 -methyladenosine (m6A) is one of the most abundant internal RNA modifications. We investigated the role of m6A-modified circRERE in osteoarthritis (OA) and its mechanism.
Materials and methods: CircRERE and IRF2BPL were screened by microarrays. The role of m6A-modification in circRERE was examined by methylated RNA precipitation and morpholino oligo (MOs) treatment. The axis of circRERE/miR-195-5p/IRF2BPL/β-catenin was determined using flow cytometry, western blotting and immunofluorescence in human chondrocytes (HCs) and corroborated using a mouse model of destabilization of medial meniscus (DMM) with intra-articular (IA) injection of adeno-associated viruses (AAV).
Results: CircRERE was decreased in OA cartilage and chondrocytes compared with control. CircRERE downregulation was likely attributed to its increased m6A modification prone to endoribonucleolytic cleavage by YTHDF2-HRSP12-RNase P/MRP in OA chondrocytes. MOs transfection targeting HRSP12 binding motifs in circRERE partially reversed decreased circRERE expression and increased apoptosis in HCs treated with IL-1β for 6 h. CircRERE exerted chondroprotective effects by targeting miR-195-5p/IRF2BPL, thus regulating the ubiquitination and degradation of β-catenin. CircRere (mouse homologue) overexpression by IA-injection of AAV-circRere into mice attenuated the severity of DMM-induced OA, whereas AAV-miR-195a-5p or AAV-sh-Irf2bpl reduced the protective effects. The detrimental effects of AAV-sh-Irf2bpl on DMM-induced OA were substantially counteracted by ICG-001, an inhibitor of β-catenin.
Conclusions: Our study is a proof-of-concept demonstration for targeting m6A-modified circRERE and its target miR-195-5p/IRF2BPL/β-catenin as potential therapeutic strategies for OA treatment.
© 2022 The Authors. Cell Proliferation published by European Cell Proliferation Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Circ_DHRS3 positively regulates GREM1 expression by competitively targeting miR-183-5p to modulate IL-1β-administered chondrocyte proliferation, apoptosis and ECM degradation.Int Immunopharmacol. 2021 Feb;91:107293. doi: 10.1016/j.intimp.2020.107293. Epub 2020 Dec 23. Int Immunopharmacol. 2021. PMID: 33360372
-
Circular RNA-9119 protects IL-1β-treated chondrocytes from apoptosis in an osteoarthritis cell model by intercepting the microRNA-26a/PTEN axis.Life Sci. 2020 Sep 1;256:117924. doi: 10.1016/j.lfs.2020.117924. Epub 2020 Jun 7. Life Sci. 2020. PMID: 32522568
-
Hsa_circ_0134111 promotes osteoarthritis progression by regulating miR-224-5p/CCL1 interaction.Aging (Albany NY). 2021 Aug 19;13(16):20383-20394. doi: 10.18632/aging.203420. Epub 2021 Aug 19. Aging (Albany NY). 2021. PMID: 34413269 Free PMC article.
-
CircSERPINE2 weakens IL-1β-caused apoptosis and extracellular matrix degradation of chondrocytes by regulating miR-495/TGFBR2 axis.Biosci Rep. 2020 Nov 27;40(11):BSR20201601. doi: 10.1042/BSR20201601. Biosci Rep. 2020. PMID: 33094798 Free PMC article.
-
Beta-catenin, cartilage, and osteoarthritis.Ann N Y Acad Sci. 2010 Mar;1192(1):344-50. doi: 10.1111/j.1749-6632.2009.05212.x. Ann N Y Acad Sci. 2010. PMID: 20392258 Free PMC article. Review.
Cited by
-
Hsa_circ_0007292 promotes chondrocyte injury in osteoarthritis via targeting the miR-1179/HMGB1 axis.J Orthop Surg Res. 2023 Jul 29;18(1):544. doi: 10.1186/s13018-023-04026-7. J Orthop Surg Res. 2023. PMID: 37516834 Free PMC article.
-
RNA modifications and their role in gene expression.Front Mol Biosci. 2025 Apr 25;12:1537861. doi: 10.3389/fmolb.2025.1537861. eCollection 2025. Front Mol Biosci. 2025. PMID: 40351534 Free PMC article. Review.
-
Novel Insights into the Regulatory Role of N6-Methyladenosine in the Pathogenesis and Clinical Treatment of Osteoarthritis: Research Status and Prospect.J Inflamm Res. 2025 May 27;18:6749-6766. doi: 10.2147/JIR.S508973. eCollection 2025. J Inflamm Res. 2025. PMID: 40453975 Free PMC article. Review.
-
iTRAQ proteomic analysis of exosomes derived from synovial fluid reveals disease patterns and potential biomarkers of Osteoarthritis.J Orthop Surg Res. 2024 Dec 19;19(1):849. doi: 10.1186/s13018-024-05336-0. J Orthop Surg Res. 2024. PMID: 39702169 Free PMC article.
-
RNA Modifications in Osteoarthritis: Epitranscriptomic Insights into Pathogenesis and Therapeutic Targets.Int J Mol Sci. 2025 May 21;26(10):4955. doi: 10.3390/ijms26104955. Int J Mol Sci. 2025. PMID: 40430096 Free PMC article. Review.
References
-
- Choi WS, Lee G, Song WH, et al. The CH25H‐CYP7B1‐RORalpha axis of cholesterol metabolism regulates osteoarthritis. Nature. 2019;566(7743):254‐258. - PubMed
-
- Hugle T, Geurts J. What drives osteoarthritis?‐synovial versus subchondral bone pathology. Rheumatology (Oxford). 2017;56(9):1461‐1471. - PubMed
-
- Martel‐Pelletier J, Barr AJ, Cicuttini FM, et al. Osteoarthritis. Nat Rev Dis Primers. 2016;2:16072. - PubMed
-
- Zheng L, Zhang Z, Sheng P, Mobasheri A. The role of metabolism in chondrocyte dysfunction and the progression of osteoarthritis. Ageing Res Rev. 2021;66:101249. - PubMed
-
- Rice SJ, Beier F, Young DA, Loughlin J. Interplay between genetics and epigenetics in osteoarthritis. Nat Rev Rheumatol. 2020;16(5):268‐281. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

