Harnessing natural-product-inspired combinatorial chemistry and computation-guided synthesis to develop N-glycan modulators as anticancer agents
- PMID: 35733906
- PMCID: PMC9159088
- DOI: 10.1039/d1sc05894k
Harnessing natural-product-inspired combinatorial chemistry and computation-guided synthesis to develop N-glycan modulators as anticancer agents
Abstract
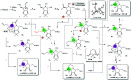
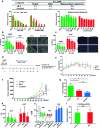
Modulation of N-glycosylation using human Golgi α-mannosidase II (α-hGMII) inhibitors is a potential anticancer approach, but the clinical utility of current α-hGMII inhibitors is limited by their co-inhibition of human lysosomal α-mannosidase (α-hLM), resulting in abnormal storage of oligomannoses. We describe the synthesis and screening of a small library of novel bicyclic iminosugar-based scaffolds, prepared via natural product-inspired combinatorial chemistry (NPICC), which resulted in the identification of a primary α-hGMII inhibitor with 13.5-fold selectivity over α-hLM. Derivatization of this primary inhibitor using computation-guided synthesis (CGS) yielded an advanced α-hGMII inhibitor with nanomolar potency and 106-fold selectivity over α-hLM. In vitro studies demonstrated its N-glycan modulation and inhibitory effect on hepatocellular carcinoma (HCC) cells. In vivo studies confirmed its encouraging anti-HCC activity, without evidence of oligomannose accumulation.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




Similar articles
-
α-D-mannose derivatives as models designed for selective inhibition of Golgi α-mannosidase II.Eur J Med Chem. 2011 Mar;46(3):944-52. doi: 10.1016/j.ejmech.2011.01.012. Epub 2011 Jan 15. Eur J Med Chem. 2011. PMID: 21295890
-
Golgi alpha-mannosidase II deficiency in vertebrate systems: implications for asparagine-linked oligosaccharide processing in mammals.Biochim Biophys Acta. 2002 Dec 19;1573(3):225-35. doi: 10.1016/s0304-4165(02)00388-4. Biochim Biophys Acta. 2002. PMID: 12417404 Review.
-
Human lysosomal alpha-mannosidases exhibit different inhibition and metal binding properties.Protein Sci. 2009 Nov;18(11):2242-51. doi: 10.1002/pro.235. Protein Sci. 2009. PMID: 19722277 Free PMC article.
-
From natural product-inspired pyrrolidine scaffolds to the development of new human Golgi α-mannosidase II inhibitors.Chem Asian J. 2013 Nov;8(11):2600-4. doi: 10.1002/asia.201300680. Epub 2013 Aug 13. Chem Asian J. 2013. PMID: 23943613 No abstract available.
-
Natural product-inspired synthesis of thiazolidine and thiazolidinone compounds and their anticancer activities.Curr Pharm Des. 2010 Jun;16(16):1826-42. doi: 10.2174/138161210791208983. Curr Pharm Des. 2010. PMID: 20337578 Review.
Cited by
-
Molecular Basis for Inhibition of Heparanases and β-Glucuronidases by Siastatin B.J Am Chem Soc. 2024 Jan 10;146(1):125-133. doi: 10.1021/jacs.3c04162. Epub 2023 Dec 20. J Am Chem Soc. 2024. PMID: 38118176 Free PMC article.
-
Synthesis, α-mannosidase inhibition studies and molecular modeling of 1,4-imino-ᴅ-lyxitols and their C-5-altered N-arylalkyl derivatives.Beilstein J Org Chem. 2023 Mar 6;19:282-293. doi: 10.3762/bjoc.19.24. eCollection 2023. Beilstein J Org Chem. 2023. PMID: 36925565 Free PMC article.
-
Mechanistic Insights into Dibasic Iminosugars as pH-Selective Pharmacological Chaperones to Stabilize Human α-Galactosidase.JACS Au. 2024 Feb 23;4(3):908-918. doi: 10.1021/jacsau.3c00684. eCollection 2024 Mar 25. JACS Au. 2024. PMID: 38559739 Free PMC article.
-
A practical synthesis of nitrone-derived C5a-functionalized isofagomines as protein stabilizers to treat Gaucher disease.Commun Chem. 2024 Apr 20;7(1):91. doi: 10.1038/s42004-024-01164-9. Commun Chem. 2024. PMID: 38643239 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

