N7-Methylguanosine Genes Related Prognostic Biomarker in Hepatocellular Carcinoma
- PMID: 35734429
- PMCID: PMC9207530
- DOI: 10.3389/fgene.2022.918983
N7-Methylguanosine Genes Related Prognostic Biomarker in Hepatocellular Carcinoma
Abstract
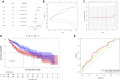
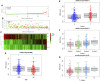
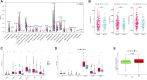
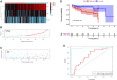
Background: About 90% of liver cancer-related deaths are caused by hepatocellular carcinoma (HCC). N7-methylguanosine (m7G) modification is associated with the biological process and regulation of various diseases. To the best of our knowledge, its role in the pathogenesis and prognosis of HCC has not been thoroughly investigated. Aim: To identify N7-methylguanosine (m7G) related prognostic biomarkers in HCC. Furthermore, we also studied the association of m7G-related prognostic gene signature with immune infiltration in HCC. Methods: The TCGA datasets were used as a training and GEO dataset "GSE76427" for validation of the results. Statistical analyses were performed using the R statistical software version 4.1.2. Results: Functional enrichment analysis identified some pathogenesis related to HCC. We identified 3 m7G-related genes (CDK1, ANO1, and PDGFRA) as prognostic biomarkers for HCC. A risk score was calculated from these 3 prognostic m7G-related genes which showed the high-risk group had a significantly poorer prognosis than the low-risk group in both training and validation datasets. The 3- and 5-years overall survival was predicted better with the risk score than the ideal model in the entire cohort in the predictive nomogram. Furthermore, immune checkpoint genes like CTLA4, HAVCR2, LAG3, and TIGT were expressed significantly higher in the high-risk group and the chemotherapy sensitivity analysis showed that the high-risk groups were responsive to sorafenib treatment. Conclusion: These 3 m7G genes related signature model can be used as prognostic biomarkers in HCC and a guide for immunotherapy and chemotherapy response. Future clinical study on this biomarker model is required to verify its clinical implications.
Keywords: hepatocellular carcinoma; m7G-related genes; nomogram; prognosis; risk-score.
Copyright © 2022 Regmi, He, Lia, Paudyal and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

