Effects of Coronary Artery Disease-Associated Variants on Vascular Smooth Muscle Cells
- PMID: 35735005
- PMCID: PMC9484647
- DOI: 10.1161/CIRCULATIONAHA.121.058389
Effects of Coronary Artery Disease-Associated Variants on Vascular Smooth Muscle Cells
Abstract
Background: Genome-wide association studies have identified many genetic loci that are robustly associated with coronary artery disease (CAD). However, the underlying biological mechanisms are still unknown for most of these loci, hindering the progress to medical translation. Evidence suggests that the genetic influence on CAD susceptibility may act partly through vascular smooth muscle cells (VSMCs).
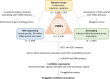
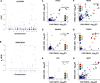
Methods: We undertook genotyping, RNA sequencing, and cell behavior assays on a large bank of VSMCs (n>1499). Expression quantitative trait locus and splicing quantitative trait locus analyses were performed to identify genes with an expression that was influenced by CAD-associated variants. To identify candidate causal genes for CAD, we ascertained colocalizations of VSMC expression quantitative trait locus signals with CAD association signals by performing causal variants identification in associated regions analysis and the summary data-based mendelian randomization test. Druggability analysis was then performed on the candidate causal genes. CAD risk variants were tested for associations with VSMC proliferation, migration, and apoptosis. Collective effects of multiple CAD-associated variants on VSMC behavior were estimated by polygenic scores.
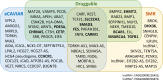
Results: Approximately 60% of the known CAD-associated variants showed statistically significant expression quantitative trait locus or splicing quantitative trait locus effects in VSMCs. Colocalization analyses identified 84 genes with expression quantitative trait locus signals that significantly colocalized with CAD association signals, identifying them as candidate causal genes. Druggability analysis indicated that 38 of the candidate causal genes were druggable, and 13 had evidence of drug-gene interactions. Of the CAD-associated variants tested, 139 showed suggestive associations with VSMC proliferation, migration, or apoptosis. A polygenic score model explained up to 5.94% of variation in several VSMC behavior parameters, consistent with polygenic influences on VSMC behavior.
Conclusions: This comprehensive analysis shows that a large percentage of CAD loci can modulate gene expression in VSMCs and influence VSMC behavior. Several candidate causal genes identified are likely to be druggable and thus represent potential therapeutic targets.
Keywords: coronary artery disease; genetics; muscle, smooth, vascular; transcriptomes.
Figures




Comment in
-
Bridging the Gap to Translating Genome-Wide Discoveries Into Therapies to Prevent and Treat Atherosclerotic Cardiovascular Disease.Circulation. 2022 Sep 20;146(12):930-933. doi: 10.1161/CIRCULATIONAHA.122.060998. Epub 2022 Sep 19. Circulation. 2022. PMID: 36121912 No abstract available.
References
-
- Erdmann J, Kessler T, Munoz Venegas L, Schunkert HA. Decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. 2018;114:1241–1257. doi: 10.1093/cvr/cvy084 - PubMed
-
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, McMahon A, Morales J, Mountjoy E, Sollis E, et al. . The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120 - PMC - PubMed
-
- Angelakopoulou A, Shah T, Sofat R, Shah S, Berry DJ, Cooper J, Palmen J, Tzoulaki I, Wong A, Jefferis BJ, et al. . Comparative analysis of genome-wide association studies signals for lipids, diabetes, and coronary heart disease: cardiovascular biomarker genetics collaboration. Eur Heart J. 2012;33:393–407. doi: 10.1093/eurheartj/ehr225 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

