Mapping Phenotypic Plasticity upon the Cancer Cell State Landscape Using Manifold Learning
- PMID: 35736000
- PMCID: PMC9353259
- DOI: 10.1158/2159-8290.CD-21-0282
Mapping Phenotypic Plasticity upon the Cancer Cell State Landscape Using Manifold Learning
Abstract
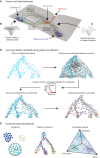
Abstract: Phenotypic plasticity describes the ability of cancer cells to undergo dynamic, nongenetic cell state changes that amplify cancer heterogeneity to promote metastasis and therapy evasion. Thus, cancer cells occupy a continuous spectrum of phenotypic states connected by trajectories defining dynamic transitions upon a cancer cell state landscape. With technologies proliferating to systematically record molecular mechanisms at single-cell resolution, we illuminate manifold learning techniques as emerging computational tools to effectively model cell state dynamics in a way that mimics our understanding of the cell state landscape. We anticipate that "state-gating" therapies targeting phenotypic plasticity will limit cancer heterogeneity, metastasis, and therapy resistance.
Significance: Nongenetic mechanisms underlying phenotypic plasticity have emerged as significant drivers of tumor heterogeneity, metastasis, and therapy resistance. Herein, we discuss new experimental and computational techniques to define phenotypic plasticity as a scaffold to guide accelerated progress in uncovering new vulnerabilities for therapeutic exploitation.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures

References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. - PubMed
-
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014;14:275–91. - PubMed
-
- Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 2012;4:127ps10. - PubMed
-
- Marine JC, Dawson SJ, Dawson MA. Non-genetic mechanisms of therapeutic resistance in cancer. Nat Rev Cancer 2020;20:743–56. - PubMed
-
- McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

