The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes
- PMID: 35736057
- PMCID: PMC9225368
- DOI: 10.3390/jof8060574
The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes
Abstract
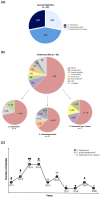
Although considered rare, the emergent Candida haemulonii species complex, formed by C. haemulonii sensu stricto (Ch), C. duobushaemulonii (Cd) and C. haemulonii var. vulnera (Chv), is highlighted due to its profile of increased resistance to the available antifungal drugs. In the present work, 25 clinical isolates, recovered from human infections during 2011-2020 and biochemically identified by automated system as C. haemulonii, were initially assessed by molecular methods (amplification and sequencing of ITS1-5.8S-ITS2 gene) for precise species identification. Subsequently, the antifungal susceptibility of planktonic cells, biofilm formation and susceptibility of biofilms to antifungal drugs and the secretion of key molecules, such as hydrolytic enzymes, hemolysins and siderophores, were evaluated by classical methodologies. Our results revealed that 7 (28%) isolates were molecularly identified as Ch, 7 (28%) as Chv and 11 (44%) as Cd. Sixteen (64%) fungal isolates were recovered from blood. Regarding the antifungal susceptibility test, the planktonic cells were resistant to (i) fluconazole (100% of Ch and Chv, and 72.7% of Cd isolates), itraconazole and voriconazole (85.7% of Ch and Chv, and 72.7% of Cd isolates); (ii) no breakpoints were defined for posaconazole, but high MICs were observed for 85.7% of Ch and Chv, and 72.7% of Cd isolates; (iii) all isolates were resistant to amphotericin B; and (iv) all isolates were susceptible to echinocandins (except for one isolate of Cd) and to flucytosine (except for two isolates of Cd). Biofilm is a well-known virulence and resistant structure in Candida species, including the C. haemulonii complex. Herein, we showed that all isolates were able to form viable biofilms over a polystyrene surface. Moreover, the mature biofilms formed by the C. haemulonii species complex presented a higher antifungal-resistant profile than their planktonic counterparts. Secreted molecules associated with virulence were also detected in our fungal collection: 100% of the isolates yielded aspartic proteases, hemolysins and siderophores as well as phospholipase (92%), esterase (80%), phytase (80%), and caseinase (76%) activities. Our results reinforce the multidrug resistance profile of the C. haemulonii species complex, including Brazilian clinical isolates, as well as their ability to produce important virulence attributes such as biofilms and different classes of hydrolytic enzymes, hemolysins and siderophores, which typically present a strain-dependent profile.
Keywords: Candida haemulonii complex; antifungal resistance; biofilm formation; hemolysins; hydrolytic enzymes; siderophores.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Cendejas-Bueno E., Kolecka A., Alastruey-Izquierdo A., Theelen B., Groenewald M., Kostrzewa M., Cuenca-Estrella M., Gomez-Lopez A., Boekhout T. Reclassification of the Candida haemulonii Complex as Candida haemulonii (C. haemulonii Group I), C. duobushaemulonii Sp. Nov. (C. haemulonii Group II), and C. haemulonii var. vulnera var. Nov.: Three Multiresistant Human Pathogenic Yeasts. J. Clin. Microbiol. 2012;50:3641–3651. doi: 10.1128/Jcm.02248-12. - DOI - PMC - PubMed
-
- Ramos L.S., Figueiredo-Carvalho M.H., Barbedo L.S., Ziccardi M., Chaves A.L., Zancope-Oliveira R.M., Pinto M.R., Sgarbi D.B., Dornelas-Ribeiro M., Branquinha M.H., et al. Candida haemulonii Complex: Species Identification and Antifungal Susceptibility Profiles of Clinical Isolates from Brazil. J. Antimicrob. Chemother. 2015;70:111–115. doi: 10.1093/jac/dku321. - DOI - PubMed
-
- Silva L.N., Oliveira S.S.C., Magalhães L.B., Andrade Neto V.V., Torres-Santos E.C., Carvalho M.D.C., Pereira M.D., Branquinha M.H., Santos A.L.S. Unmasking the Amphotericin B Resistance Mechanisms in Candida haemulonii Species Complex. ACS Infect. Dis. 2020;6:1273–1282. doi: 10.1021/acsinfecdis.0c00117. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

