Transcription Factor Mavib-1 Negatively Regulates Conidiation by Affecting Utilization of Carbon and Nitrogen Source in Metarhizium acridum
- PMID: 35736077
- PMCID: PMC9224900
- DOI: 10.3390/jof8060594
Transcription Factor Mavib-1 Negatively Regulates Conidiation by Affecting Utilization of Carbon and Nitrogen Source in Metarhizium acridum
Abstract
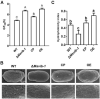
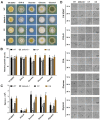
Conidium is the main infection unit and reproductive unit of pathogenic fungi. Exploring the mechanism of conidiation and its regulation contributes to understanding the pathogenicity of pathogenic fungi. Vib-1, a transcription factor, was reported to participate in the conidiation process. However, the regulation mechanism of Vib-1 in conidiation is still unclear. In this study, we analyzed the function of Vib-1 and its regulation mechanism in conidiation through knocking out and overexpression of Vib-1 in entomopathogenic fungus Metarhizium acridum. Results showed that the colonial growth of Mavib-1 disruption mutant (ΔMavib-1) was significantly decreased, and conidiation was earlier compared to wild type (WT), while overexpression of Mavib-1 led to a delayed conidiation especially when carbon or nitrogen sources were insufficient. Overexpression of Mavib-1 resulted in a conidiation pattern shift from microcycle conidiation to normal conidiation on nutrient-limited medium. These results indicated that Mavib-1 acted as a positive regulator in vegetative growth and a negative regulator in conidiation by affecting utilization of carbon and nitrogen sources in M. acridum. Transcription profile analysis demonstrated that many genes related to carbon and nitrogen source metabolisms were differentially expressed in ΔMavib-1 and OE strains compared to WT. Moreover, Mavib-1 affects the conidial germination, tolerance to UV-B and heat stresses, cell wall integrity, conidial surface morphology and conidial hydrophobicity in M. acridum. These findings unravel the regulatory mechanism of Mavib-1 in fungal growth and conidiation, and enrich the knowledge to conidiation pattern shift of filamentous fungi.
Keywords: carbon and nitrogen utilization; conidiation; entomopathogenic fungi; transcription factor Mavib-1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Zhang S.Z., Peng G.X., Xia Y.X. Microcycle conidiation and the conidial properties in the entomopathogenic fungus Metarhizium acridum on agar medium. Biocontrol Sci. Technol. 2010;20:809–819. doi: 10.1080/09583157.2010.482201. - DOI
-
- De Faria M.R., Wraight S.P. Mycoinsecticides and mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control. 2007;43:237–256. doi: 10.1016/j.biocontrol.2007.08.001. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

