Post-Translational Modifications of PCNA: Guiding for the Best DNA Damage Tolerance Choice
- PMID: 35736104
- PMCID: PMC9225081
- DOI: 10.3390/jof8060621
Post-Translational Modifications of PCNA: Guiding for the Best DNA Damage Tolerance Choice
Abstract
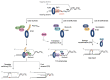
The sliding clamp PCNA is a multifunctional homotrimer mainly linked to DNA replication. During this process, cells must ensure an accurate and complete genome replication when constantly challenged by the presence of DNA lesions. Post-translational modifications of PCNA play a crucial role in channeling DNA damage tolerance (DDT) and repair mechanisms to bypass unrepaired lesions and promote optimal fork replication restart. PCNA ubiquitination processes trigger the following two main DDT sub-pathways: Rad6/Rad18-dependent PCNA monoubiquitination and Ubc13-Mms2/Rad5-mediated PCNA polyubiquitination, promoting error-prone translation synthesis (TLS) or error-free template switch (TS) pathways, respectively. However, the fork protection mechanism leading to TS during fork reversal is still poorly understood. In contrast, PCNA sumoylation impedes the homologous recombination (HR)-mediated salvage recombination (SR) repair pathway. Focusing on Saccharomyces cerevisiae budding yeast, we summarized PCNA related-DDT and repair mechanisms that coordinately sustain genome stability and cell survival. In addition, we compared PCNA sequences from various fungal pathogens, considering recent advances in structural features. Importantly, the identification of PCNA epitopes may lead to potential fungal targets for antifungal drug development.
Keywords: DNA damage tolerance; DNA replication forks; DNA replication stress; PCNA; fungal genome stability; post-translational modifications; salvage recombination; template switch; translesion synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
-
DNA-damage tolerance through PCNA ubiquitination and sumoylation.Biochem J. 2020 Jul 31;477(14):2655-2677. doi: 10.1042/BCJ20190579. Biochem J. 2020. PMID: 32726436 Review.
-
Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA.Cell Res. 2008 Jan;18(1):162-73. doi: 10.1038/cr.2007.114. Cell Res. 2008. PMID: 18157158 Review.
-
The roles of PCNA SUMOylation, Mms2-Ubc13 and Rad5 in translesion DNA synthesis in Saccharomyces cerevisiae.Mol Microbiol. 2011 May;80(3):786-97. doi: 10.1111/j.1365-2958.2011.07610.x. Epub 2011 Mar 16. Mol Microbiol. 2011. PMID: 21362066
-
DNA Damage Tolerance Pathway Choice Through Uls1 Modulation of Srs2 SUMOylation in Saccharomyces cerevisiae.Genetics. 2017 May;206(1):513-525. doi: 10.1534/genetics.116.196568. Epub 2017 Mar 24. Genetics. 2017. PMID: 28341648 Free PMC article.
Cited by
-
Molecular mechanism of PARP inhibitor resistance.Oncoscience. 2024 Sep 23;11:69-91. doi: 10.18632/oncoscience.610. eCollection 2024. Oncoscience. 2024. PMID: 39318358 Free PMC article. Review.
-
Isorhamnetin alleviates symptoms and inhibits oxidative stress levels in rats with pulmonary arterial hypertension.Iran J Basic Med Sci. 2024;27(12):1616-1623. doi: 10.22038/ijbms.2024.75860.16421. Iran J Basic Med Sci. 2024. PMID: 39539446 Free PMC article.
-
Echinatin inhibits tumor growth and synergizes with chemotherapeutic agents against human bladder cancer cells by activating p38 and suppressing Wnt/β-catenin pathways.Genes Dis. 2023 May 18;11(2):1050-1065. doi: 10.1016/j.gendis.2023.03.031. eCollection 2024 Mar. Genes Dis. 2023. PMID: 37692489 Free PMC article.
-
Effects of Defective Unloading and Recycling of PCNA Revealed by the Analysis of ELG1 Mutants.Int J Mol Sci. 2023 Jan 13;24(2):1568. doi: 10.3390/ijms24021568. Int J Mol Sci. 2023. PMID: 36675081 Free PMC article.
-
From Processivity to Genome Maintenance: The Many Roles of Sliding Clamps.Genes (Basel). 2022 Nov 7;13(11):2058. doi: 10.3390/genes13112058. Genes (Basel). 2022. PMID: 36360296 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

