Characterizing SARS-CoV-2 Viral Clearance Kinetics to Improve the Design of Antiviral Pharmacometric Studies
- PMID: 35736134
- PMCID: PMC9295592
- DOI: 10.1128/aac.00192-22
Characterizing SARS-CoV-2 Viral Clearance Kinetics to Improve the Design of Antiviral Pharmacometric Studies
Abstract
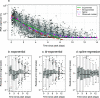
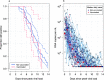
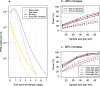
A consensus methodology for the pharmacometric assessment of candidate SARS-CoV-2 antiviral drugs would be useful for comparing trial results and improving trial design. The time to viral clearance, assessed by serial qPCR of nasopharyngeal swab samples, has been the most widely reported measure of virological response in clinical trials, but it has not been compared formally with other metrics, notably model-based estimates of the rate of viral clearance. We analyzed prospectively gathered viral clearance profiles from 280 infection episodes in vaccinated and unvaccinated individuals. We fitted different phenomenological pharmacodynamic models (single exponential decay, bi-exponential, penalized splines) and found that the clearance rate, estimated from a mixed effects single exponential decay model, is a robust pharmacodynamic summary of viral clearance. The rate of viral clearance, estimated from viral densities during the first week following peak viral load, provides increased statistical power (reduced type 2 error) compared with time to clearance. Antiviral effects approximately equivalent to those with currently used and recommended SARS-CoV-2 antiviral treatments, notably nirmatrelvir and molnupiravir, can be detected from randomized trials with sample sizes of only 35 to 65 patients per arm. We recommend that pharmacometric antiviral assessments should be conducted in early COVID-19 illness with serial qPCR samples taken over 1 week.
Keywords: SARS-CoV-2; antiviral drugs; clearance rate; pharmacodynamics; phase 2; time to clearance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- He X, Lau EH, Wu P, Deng X, Wang J, Hao X, Lau YC, Wong JY, Guan Y, Tan X, Mo X, Yanqing C, Baolin L, Chen W, Hu F, Zhang Q, Zhong M, Wu Y, Zhao L, Zhang F, Cowling BJ, Li F, Leung G. 2020. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med 26:672–675. 10.1038/s41591-020-0869-5. - DOI - PubMed
-
- Kissler SM, Fauver JR, Mack C, Tai CG, Breban MI, Watkins AE, Samant RM, Anderson DJ, Metti J, Khullar G, Baits R, MacKay M, Salgado D, Baker T, Dudley JT, Mason CE, Ho DD, Grubaugh ND, Grad YH. 2021. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med 385:2489–2491. 10.1056/NEJMc2102507. - DOI - PMC - PubMed
-
- Salje H, Kiem CT, Lefrancq N, Courtejoie N, Bosetti P, Paireau J, Andronico A, Hozé N, Richet J, Dubost C-L, Strat YL, Lessler J, Levy-Bruhl D, Fontanet A, Opatowski L, Boelle P-Y, Cauchemez S. 2020. Estimating the burden of SARS-CoV-2 in France. Science 369:208–211. 10.1126/science.abc3517. - DOI - PMC - PubMed
-
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, Bailey R, Swanson KA, Roychoudhury S, Koury K, Li P, Kalina WV, Cooper D, Frenck RW, Hammitt LL, Tureci O, Nell H, Schaefer A, Ünal S, Tresnan DB, Mather S, Dormitzer PR, Şahin U, Jansen KU, Gruber WC, C4591001 Clinical Trial Group . 2020. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med 383:2603–2615. 10.1056/NEJMoa2034577. - DOI - PMC - PubMed
-
- Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators . 2021. REGN-COV2, a neutralizing antibody cocktail, in outpatients with COVID-19. N Engl J Med 384:238–251. 10.1056/NEJMoa2035002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

